Background: Temperature concomitantly modulates kinetic and adsorption properties in heterogeneous enzyme catalysis.
Results: Affinity-activity relationships for four Cel7A cellobiohydrolases are characterized over a broad temperature interval.
Conclusion: Cellobiohydrolases are strongly activated by temperature at high, but not at low, substrate loads.
Significance: Fundamental insight into cellulolytic mechanisms at high (industrially relevant) temperatures is gained.
Keywords: carbohydrate-binding protein, cellulase, enzyme kinetics, Hypocrea jecorina, thermophile, Rasamsonia emersonii, thermoactivation
Abstract
We measured hydrolytic rates of four purified cellulases in small increments of temperature (10–50 °C) and substrate loads (0–100 g/liter) and analyzed the data by a steady state kinetic model that accounts for the processive mechanism. We used wild type cellobiohydrolases (Cel7A) from mesophilic Hypocrea jecorina and thermophilic Rasamsonia emersonii and two variants of these enzymes designed to elucidate the role of the carbohydrate binding module (CBM). We consistently found that the maximal rate increased strongly with temperature, whereas the affinity for the insoluble substrate decreased, and as a result, the effect of temperature depended strongly on the substrate load. Thus, temperature had little or no effect on the hydrolytic rate in dilute substrate suspensions, whereas strong temperature activation (Q10 values up to 2.6) was observed at saturating substrate loads. The CBM had a dual effect on the activity. On one hand, it diminished the tendency of heat-induced desorption, but on the other hand, it had a pronounced negative effect on the maximal rate, which was 2-fold larger in variants without CBM throughout the investigated temperature range. We conclude that although the CBM is beneficial for affinity it slows down the catalytic process. Cel7A from the thermophilic organism was moderately more activated by temperature than the mesophilic analog. This is in accord with general theories on enzyme temperature adaptation and possibly relevant information for the selection of technical cellulases.
Introduction
Cel7 cellobiohydrolases (cellulose 1,4-β-cellobiosidase (reducing end): EC 3.2.1.176) are among the most effective cellulolytic enzymes and play an essential role in decomposition processes performed for example by ascomycete fungi. They also make up the dominant component in enzyme mixtures used industrially to convert lignocellulosic biomass to fermentable sugars (so-called saccharification). Both of these aspects have generated a substantial research interest in this group of enzymes (1, 2), and many structural, mechanistic, and phylogenic questions have been elucidated over the past decades. This work has established that some cellobiohydrolases follow a quite extraordinary reaction path with an initial attack on the end of the cellulose strand followed by sequential release of cellobiose as the enzyme slides along the cellulose crystal with a single polysaccharide strand threaded through a long tunnel with the catalytic site located toward the end. This processive mechanism is considered an effective way (3–5) to overcome the high chemical and physical stability of cellulose (6, 7) and therefore an important element in natural carbon cycling.
One key question in both fundamental and applied Cel7A research is how temperature affects this complex process, and different aspects of this have been addressed in earlier studies. Many of these have been motivated by industrial questions or aimed at clarifying the optimal temperature range for different wild type enzymes or engineered variants with improved thermal stability (8–13). This type of work typically quantifies the amount of soluble sugar produced from an insoluble substrate in end point measurements at variable temperatures with other experimental parameters kept constant. Some studies have applied a broader methodology and investigated temperature effects as a function of contact time and the loads of enzyme and (insoluble) substrate, respectively, and used this to identify optimal hydrolysis conditions (14, 15). In many cases, the overall rate of cellulose hydrolysis for purified enzyme or enzyme mixtures has been measured at different temperatures and interpreted along the lines of the Arrhenius equation. However, reported activation energies, Ea, vary (16–22), and in some cases, it is difficult to assign a physical meaning to reported Ea values as they are not defined with respect to a specific reaction scheme. Some studies have analyzed temperature effects on cellulolytic activity with respect to specific kinetic models. The majority of this work has used soluble substrate analogs, which are convenient to assay and allow interpretation within the Michaelis-Menten framework (1, 23, 24). This approach has provided important knowledge of the temperature response of cellulases including insight into differences between enzymes from organisms adapted to different temperatures. In contrast, work on small soluble substrates obviously does not capture any special behavior pertaining to processive hydrolysis of an insoluble substrate. A few studies have addressed this latter issue and analyzed the results with respect to deterministic models. Examples of this includes work by Zhang et al. (16), who studied the hydrolysis of rice straw by a crude cellulase extract between 37 and 50 °C and used a fractal kinetic model to rationalize the results. More recently, Ye and Berson (26) tested a kinetic model that accounted for both hydrolysis and enzyme inactivation against experimental data for a commercial enzyme mixture at different temperatures. Their results suggested that Ea for hydrolysis and inactivation was of comparable size (∼70 kJ/mol). Brown et al. (27) tested a number of previously developed steady state models against a set of data based on lignocellulosic biomass and a fungal enzyme mixture. They concluded that a three-parameter model, which accounted for the number of reactive sites covered by the enzymes, represented their data well. This approach gave a linear Arrhenius plot with an activation energy of 48 kJ/mol.
However, systematic investigations of temperature effects on the hydrolysis of insoluble substrate are scarce, and we are not aware of any earlier studies on monocomponent cellulases that report systematic temperature-activity data and rationalize them with respect to a relevant theoretical framework. To address this, we implemented a medium throughput assay and measured activity and adsorption in small increments of temperature (10–50 °C) and substrate load (0–100 g/liter). Results from 1-h trials with a pure cellulose substrate (Avicel) were used to estimate steady state reaction rates at low degrees of conversion and were analyzed with respect to a Michaelis-Menten-type model for processive enzymes described previously (28). We report the temperature dependence of kinetic parameters for four processive enzymes, which were selected to clarify affinity-activity relationships and the effect of natural adaptation to higher temperatures. Specifically, we compared Cel7A enzymes from Hypocrea jecorina (often identified by the name of its anamorph, Trichoderma reesei) and the thermophile Rasamsonia emersonii (previously Talaromyces emersonii) (29). The catalytic domains of these two enzymes are structurally homologous and have a sequence identity of 66% (30), but the intact enzymes are distinctively different in the sense that H. jecorina Cel7A has a two-module architecture with a catalytic domain and a family I carbohydrate binding module (CBM)2 connected through a flexible, glycosylated linker (31). Conversely, R. emersonii Cel7A only consists of a catalytic domain (30, 32). In addition to these two wild type enzymes, we also studied two enzyme variants designed to highlight the role of the CBM. One was the catalytic domain of H. jecorina Cel7A without CBM and linker, and the other was a chimeric protein composed of the linker and CBM from H. jecorina Cel7A and the R. emersonii enzyme. Henceforth, we will refer to these enzymes by their origin (Hj or Re) followed by subscript “CORE” or “CBM” for one- and two-domain variants, respectively (e.g. HjCBM for the H. jecorina wild type).
Experimental Procedures
Enzymes
H. jecorina Cel7A and R. emersonii Cel7A wild types were expressed in Aspergillus oryzae as described previously (33). Expression of the fusion protein (ReCBM) and the H. jecorina Cel7A core (HjCORE) (residues 18–453 of UniProt entry G0RVK1) has also been described earlier (33). All enzymes were purified from fermentation broths by hydrophobic interaction chromatography followed by ion exchange chromatography on the ÄKTA system (GE Healthcare). Fermentation broths were filtered through a bottle polyethersulfone top filter with a 0.22-μm cutoff, and ammonium sulfate was added to make a 1.8 m solution. Subsequently, the fermentation broths were applied to a 200-ml phenyl-Sepharose® 6 (high sub) FastFlow column XK50 (GE Healthcare), which had been pre-equilibrated with 1.8 m ammonium sulfate, 25 mm HEPES, pH 7.0. The column was washed with equilibration buffer followed by 0.54 m ammonium sulfate. Cel7As were batch-eluted with 25 mm HEPES, pH 7.0 and desalted on a SephadexTM G-25 (medium) column (GE Healthcare) equilibrated with 25 mm MES, pH 6.0. Next, the Cel7As were applied to a 60-ml SOURCETM 15Q column (GE Healthcare) equilibrated with 25 mm MES, pH 6.0. Cel7As from H. jecorina were eluted with a linear 50–300 mm sodium chloride gradient for 3 column volumes, whereas R. emersonii enzymes were eluted with a 100–200 mm sodium chloride gradient for 1.5 column volumes followed by 1.5 column volumes of 300 mm sodium chloride. Fractions were analyzed for the presence of Cel7A by SDS-PAGE using 12-well NuPAGE® 4–12% Bis-Tris gel (GE Healthcare). As estimated by SDS-PAGE and non-denaturing PAGE, the cellulases were purified to apparent homogeneity. The concentration of purified enzyme stocks was measured by amino acid analysis. Protein samples were dried down and hydrolyzed in 18.5% HCl, 0.1% phenol at 110 °C for 16 h. Amino acid analyses were performed by precolumn derivatization using the Waters AccQ-Tag Ultra method. In short, amino acids were derivatized by the AccQ-Tag Ultra reagent, separated with reversed phase ultraperformance LC (Waters), and the derivatives were quantitated based on UV absorbance. Enzyme concentrations were also measured by conventional UV absorption (34) at 280 nm. We used the following molar extinction coefficients: HjCBM, 86,760 m−1 cm−1; HjCORE, 80,550 m−1 cm−1; ReCBM, 81,135 m−1 cm−1; and ReCORE, 74,925 m−1 cm−1. No systematic differences between these two methods were detected.
Activity Assay
Hydrolysis of insoluble cellulose was quantified by the para-hydroxybenzoic acid hydrazide method (35). Avicel PH101 (Sigma-Aldrich) was washed six times in Milli-Q water and twice in buffer (50 mm acetate, pH 5.0; henceforth called standard buffer). Washed Avicel PH101 was suspended in 50 mm acetate, pH 5.0, and aliquots of 230 μl with loads between 0 and 106 g/liter were transferred to 96-well plates (96F 26960, Thermo Scientific). The outermost wells of the microtiter plate were not used for hydrolysis experiments. The plates were initially equilibrated at the experimental temperature (10–50 °C) for 20 min in an Eppendorf Thermomixer, and the reaction was started by the addition of 20 μl of enzyme stocks to a final concentration of 400 nm. The plates were mixed at 1100 rpm at the desired temperature for 1 h, and the reaction was then stopped by centrifugation for 3 min at 3500 rpm at 5 °C (Hereaus Multifuge 3 S-R). The choice of reaction time and enzyme concentration was made after initial trials had shown readily detectable product concentrations (>∼1 μm) at the lowest temperatures and substrate loads studied here. It was also suitable in the sense that product concentrations in samples with the highest substrate loads were much lower than published inhibition constants for Cel7A (36–38). Hence, we neglected product inhibition in the data analysis. To measure the concentration of soluble, reducing sugars, 50 μl of supernatant was transferred to 96-well PCR sample tubes (0.2-ml non-skirted 96-well PCR plate, AB0600, Thermo Scientific) and 75 μl of 15 mg/ml para-hydroxybenzoic acid hydrazide solution (4-hydroxybenzhydrazide; H9882, Sigma) dissolved in buffer (0.18 m potassium sodium tartrate (108087, Merck) was added. Subsequently, the PCR sample tubes were placed in a Peltier thermal cycler and incubated at 95 °C for 10 min and 20 °C for 5 min. One hundred microliters were transferred from the 96-well PCR sample tubes to a 96-well plate (96F 26960, Thermo Scientific). Absorption at 405 nm was determined in a plate reader (Molecular Devices SpectraMax M2), and the soluble reducing sugars were quantified based on standards with 0–0.5 mm cellobiose. Blanks (without enzyme) were included and subtracted for all measurements. All experiments were carried out in triplicates.
Adsorption
One hundred microliters of supernatant was retrieved from each sample in the activity assay and transferred to a 96-well microtiter plate (655079, Greiner Bio One). The intrinsic protein fluorescence at 340 nm was determined in a plate reader (Molecular Devices SpectraMax M2) using an excitation wavelength of 280 nm. For these experiments, standard curves ranging from 0 to 800 nm Cel7A (HjCBM, HjCORE, ReCBM, or ReCORE) were included.
Kinetic Theory
Measurements of hydrolytic activity were analyzed by a steady state model for processive enzymes (28, 39). The starting point is a simplified scheme (Scheme 1), which uses three rate constants and a processivity number to describe the reaction. It is assumed that the free enzyme, E, combines with a cellulose strand, Cm, to form a complex, ECm. The complex now goes through consecutive catalytic steps, which release the product cellobiose, C, and concomitantly shorten the cellulose strand to ECm − 1, ECm − 2, etc. This course is governed by the rate constants, pkon, pkcat, and pkoff as specified in the scheme (subscript p in front of the parameter indicates its relationship to the processive Scheme 1). The last parameter, n, is the average number of steps following one association (the processivity number), which can be measured experimentally albeit with some technical challenges (40). We note that Scheme 1 was chosen as a compromise between the structural and empirical understanding of Cel7A catalysis on one hand and simplicity on the other hand. Other descriptions of the reaction course could be equally meaningful and able to fit the data, although we surmise that the current level of structural and quantitative data for association and dissociation would be too limited to justify more complex reaction schemes.
SCHEME 1.
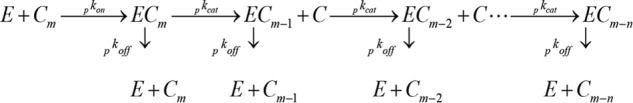
Simplified reaction scheme used to characterize processive activity of Cel7A enzymes against insoluble cellulose.
The steady state rate of cellobiose production, pvss = d[C[/dt, can be expressed (28) as follows.
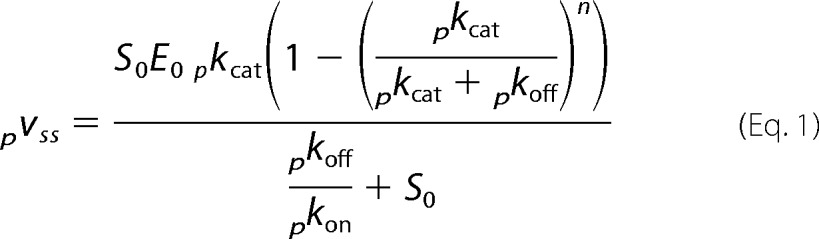 |
where E0 and S0 are the total concentration of enzyme (in μm) and substrate (in g/liter), respectively. To simplify Equation 1 and illustrate its relationship to the usual Michaelis-Menten equation, we define a maximal processive rate, pVmax, and a processive analog of the Michaelis constant, pKm.
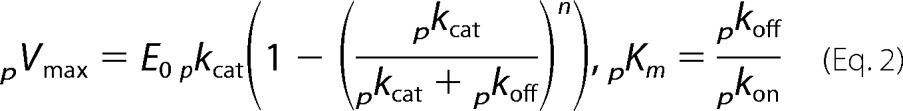 |
Inserting Equation 2 into Equation 1 yields the usual hyperbolic form.
 |
We note that pvss is an approximation of the steady state rate as Cel7A (as well as other cellulases) show so-called non-linear kinetics where the progress curves never become fully linear. However, we have previously discussed advantages and limitations of Equation 3 and suggested that it is useful for the analysis of short experiments like those considered here (28). The strategy for the current work was to fit Equation 3 to experimental data for pvss(S0) at different temperatures and for different enzymes. The resulting parameters, pVmax and pKm, and their temperature dependence were then used to illustrate temperature effects on affinity and kinetics.
Results
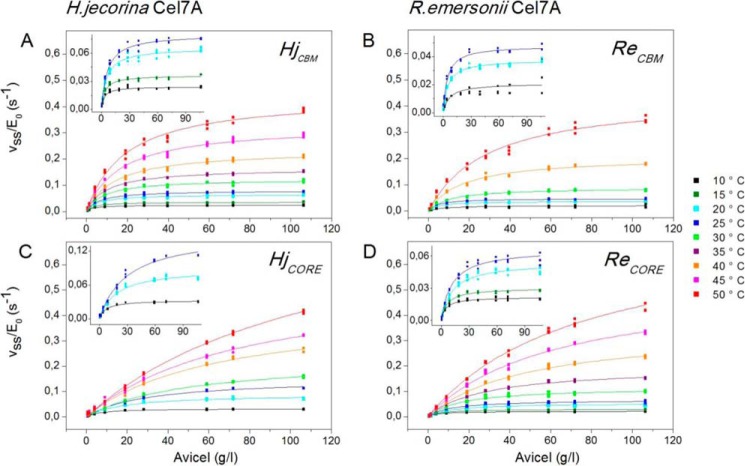
Steady state rates, pvss, for the four investigated enzymes were estimated as the ratio of the final cellobiose concentration and the contact time (1 h) and normalized with respect to the total enzyme concentration (E0 = 0.40 μm) to obtain the specific activity, pvss/E0, in units of s−1. Fig. 1 illustrates how this parameter changed with substrate load and temperature; symbols represent experimental data, and the lines are best fits of Equation 3. It appears that the model accounted well for the data under all investigated conditions. The highest conversions of Avicel reached in these experiments were below 1% (in most cases much below).
FIGURE 1.
Specific enzyme activity (pvss/E0) for HjCBM (A), ReCBM (B), HjCORE (C), and ReCORE (D) plotted as a function of Avicel load (0–106 g/liter) between 10 and 50 °C. Symbols represent all experimental data from triplicate measurements, and lines are the best fit of Equation 3. Insets show enlargements of results at the lower temperatures, which are hard to assess on the main figures.
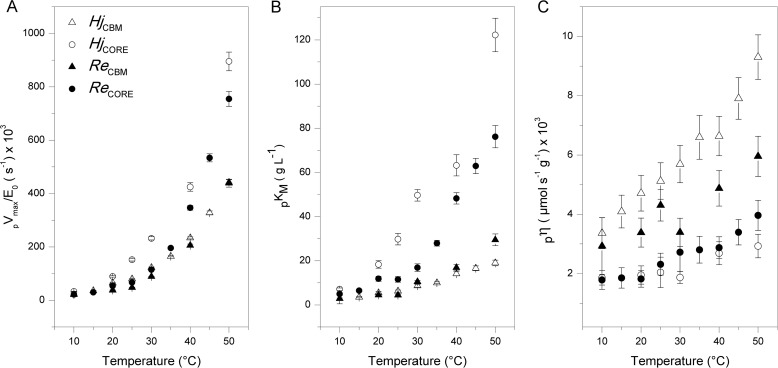
The kinetic parameters pVmax/E0 and pKm derived from the non-linear regression are listed in Table 1 and plotted as a function of temperature in Fig. 2. In most cases, the highest measured rate was close to pVmax, and as a result, the two kinetic parameters in Equation 3 could be well resolved with very low parameter interdependence. However, for one-domain enzymes at the highest investigated temperatures, saturating conditions were well above the accessible range of substrate loads. This led to parameter interdependence between pVmax and pKm and hence larger uncertainty (see Table 1). In these cases, only the initial slope (the specificity constant; see below) was precisely determined. One interesting result here is that at specified temperatures both pKm and pVmax are consistently higher for the one-domain enzymes (HjCORE and ReCORE) compared with enzymes with a CBM (HjCBM and ReCBM). These differences are quite pronounced. Hence, pVmax is about twice as high for the one-domain enzymes throughout the investigated temperature range, and the effect on the CBM on pKm is even larger. Table 1 and Fig. 2 also show the processive analog of the specificity constant, pη.
 |
TABLE 1.
Temperature dependence of kinetic and adsorption parameters for two Cel7A wild types, HjCBM and ReCORE, and the corresponding constructs HjCORE and ReCBM
Maximal specific rates, pVmax/E0 (±S.E.) and processive Michaelis constants, pKm (±S.E.) as defined in Equation 2 were derived from the regression analysis shown in Fig. 1. These two parameters were used to calculate the processive specificity constant, pη, defined in Equation 4. The last column shows the partitioning coefficient calculated from the free enzyme concentration measured at the end of the 1-h hydrolysis experiment (see Fig. 5).
| T | pVmax/E0 | pKm | pη | Kp |
|---|---|---|---|---|
| °C | s−1 × 103 | g liter−1 | (μmol s−1 g−1) × 103 | liter g−1 |
| HjCBM | ||||
| 10 | 24 ± 1 | 2.8 ± 0.3 | 3.4 ± 0.5 | 2.0 |
| 15 | 36 ± 1 | 3.5 ± 0.3 | 4.1 ± 0.6 | 2.2 |
| 20 | 65 ± 1 | 5.5 ± 0.5 | 4.7 ± 0.6 | 2.0 |
| 25 | 80 ± 1 | 6.2 ± 0.5 | 5.1 ± 0.6 | 1.5 |
| 30 | 123 ± 2 | 8.6 ± 0.6 | 5.7 ± 0.6 | 0.8 |
| 35 | 165 ± 2 | 10 ± 0.4 | 6.6 ± 0.6 | 1.0 |
| 40 | 234 ± 4 | 14 ± 0.9 | 6.6 ± 0.6 | 0.9 |
| 45 | 328 ± 7 | 17 ± 1.1 | 7.9 ± 0.6 | 0.5 |
| 50 | 442 ± 10 | 19 ± 1.3 | 9.3 ± 0.8 | 0.3 |
| HjCORE | ||||
| 10 | 33 ± 1 | 7.1 ± 0.5 | 1.9 ± 0.2 | - |
| 20 | 90 ± 3 | 18 ± 1.7 | 2.0 ± 0.3 | 0.09 |
| 25 | 152 ± 5 | 30 ± 2.6 | 2.0 ± 0.5 | 0.05 |
| 30 | 232 ± 6 | 50 ± 2.6 | 1.9 ± 0.2 | 0.05 |
| 40 | 425 ± 16 | 63 ± 4.7 | 2.7 ± 0.4 | 0.04 |
| 50 | 895 ± 35 | 122 ± 8 | 2.9 ± 0.4 | 0.02 |
| ReCBM | ||||
| 10 | 21 ± 2 | 2.8 ± 2.4 | 2.9 ± 1.0 | 2.3 |
| 20 | 38 ± 1 | 4.5 ± 0.4 | 3.4 ± 0.5 | 1.0 |
| 25 | 48 ± 1 | 4.5 ± 0.3 | 4.3 ± 0.5 | 0.5 |
| 30 | 89 ± 1 | 10 ± 0.7 | 3.4 ± 0.5 | 0.7 |
| 40 | 206 ± 5 | 17 ± 1.4 | 4.9 ± 0.6 | 0.3 |
| 50 | 439 ± 15 | 29 ± 2.7 | 5.0 ± 0.7 | 0.6 |
| ReCORE | ||||
| 10 | 22 ± 1 | 4.9 ± 0.6 | 1.8 ± 0.3 | 0.5 |
| 15 | 30 ± 1 | 6.5 ± 0.5 | 1.9 ± 0.3 | 0.4 |
| 20 | 54 ± 1 | 12 ± 1.2 | 1.8 ± 0.3 | 0.2 |
| 25 | 67 ± 2 | 12 ± 1.3 | 2.3 ± 0.4 | 0.1 |
| 30 | 115 ± 4 | 17 ± 1.7 | 2.7 ± 0.6 | 0.2 |
| 35 | 196 ± 3 | 30 ± 1.3 | 2.8 ± 0.5 | 0.1 |
| 40 | 347 ± 9 | 48 ± 2.6 | 2.9 ± 0.4 | 0.08 |
| 45 | 534 ± 15 | 63 ± 3.5 | 3.4 ± 0.4 | 0.07 |
| 50 | 755 ± 28 | 76 ± 5.1 | 4.0 ± 0.5 | 0.03 |
FIGURE 2.
Maximum specific rate (pVmax/E0; A), Michaelis constant (pKm; B), and specificity constant (pη; C) plotted as a function of temperature for the four investigated enzymes.
The temperature dependence of this parameter is illustrated in Fig. 2C, and it appears that pη is both larger and more temperature-dependent for two-domain enzymes compared with variants with no CBM.
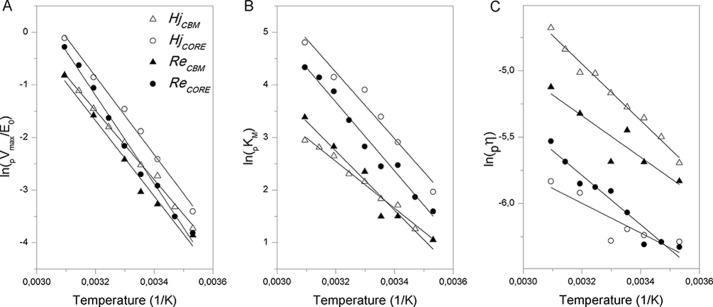
To further characterize temperature dependence, we made Arrhenius plots (i.e. natural logarithm of the parameter plotted against the reciprocal of the absolute temperature) for the data in Fig. 2. The results in Fig. 3 show linear relations, although the experimental scatter for pη was quite high in some cases. Activation energies, Ea, derived from the slopes in Fig. 3 were 60–70 kJ/mol for pVmax (Fig. 3A), 37–52 kJ/mol for pKm (Fig. 3B), and 9–16 kJ/mol for pη (Fig. 3C).
FIGURE 3.
Arrhenius plots. The natural logarithm of the maximum specific rate (pVmax; A), the Michaelis constant (pKm; B), and specificity constant (pη; C) are plotted against the reciprocal of the absolute temperature.
Implications of these energy barriers along with other activation parameters pertaining to Scheme 1 are discussed in the companion article (62). Here, we are only interested in these plots as a practical measure of the temperature dependence of a kinetic parameter. One intuitive way to express the logarithmic relation is the so-called Q10 value, which signifies the relative increment upon a 10 °C temperature increase. For many enzymes reactions around room temperature, Q10 for Vmax has been shown to be about 2, thus implying that the reaction rate at saturating substrate loads doubles upon a 10 °C temperature rise (Q10 = 2 at room temperature corresponds to an Ea of about 50 kJ/mol). The results in Fig. 3 corresponded3 to Q10 values of 2.1–2.6 for pVmax, whereas it was 1.6–2.1 for pKm and only 1.1–1.2 for pη. To illustrate the meaning of this, we first note that the specificity constant, pη, is the slope of the (near-linear) part of the curves in Fig. 1 at low substrate loads (i.e. for S ≪ pKm). It follows that pη may be interpreted as an apparent second order rate constant, which governs the kinetics at low substrate loads, vss ≈ pη[E]0[S] (41). This means that the reaction was much more activated by temperature at high substrate loads (Q10 for pVmax was 2.1–2.6) than at low substrate loads (Q10 for pη was 1.1–1.2). It is also interesting to consider average Q10 values for the investigated one- and two-domain enzymes and hence the effect of the CBM on temperature activation. For enzymes with CBM, we found Q10 values of 2.17 ± 0.10 and 1.76 ± 0.16 for pVmax and pKm, respectively. The analogous (average) values for one-domain enzymes were 2.44 ± 0.15 and 2.10 ± 0.09, and these numbers revealed that the binding module reduced the sensitivity to temperature of both pVmax and pKm.
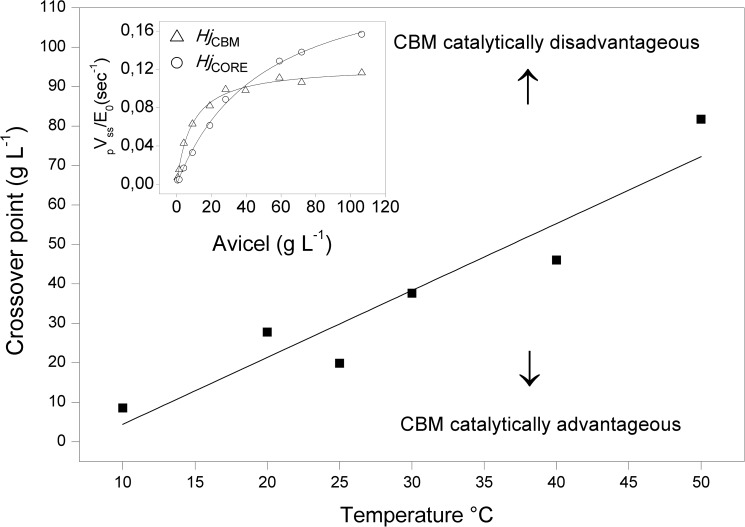
To further illustrate relationships among temperature, activity, and the CBM, we replotted data from Fig. 1 to directly compare pairs of enzymes with and without binding module. An example for H. jecorina (i.e. a comparison of HjCBM and HjCORE) at T = 30 °C is shown in the inset of Fig. 4. It appeared that the two-domain enzyme was more active at low substrate load (below ∼35 g/liter), whereas the opposite was true at higher loads. The effect was quite pronounced, and the one-domain enzyme outperformed the wild type by about 40% at the highest loads in Fig. 4. It also appears from the figure that the one-domain enzyme does not reach saturating conditions in the current experiments, and this underlies the observation (Table 1) that the difference in maximal rates for the two variants was even larger (about 2-fold) than the difference in pvss shown in Fig. 4. Analogous plots revealed that a crossover occurred at all investigated temperatures (and for both Hj and Re enzymes), and the main panel in Fig. 4 shows how the substrate load at the crossover point changed with temperature. This plot shows that at 10 °C the one-domain variant was most active except in very dilute substrate suspensions (below 5–10 g/liter). Conversely, at 50 °C, the two-domain enzyme was more effective over most of the investigated range and only became inferior to the CBM-free variant when the substrate load exceeded 70–80 g/liter.
FIGURE 4.
Effects of temperature and binding module on the activity of Cel7A from H. jecorina. The inset shows data from 30 °C of the specific rate versus substrate load. It appears that the one-domain variant was slower at low substrate but became faster than the two-domain enzyme above ∼40 g/liter. The main panel shows the location (i.e. substrate load) of this crossover as a function of temperature. The line separates the plane into regions where the CBM promoted (upper left) or reduced (lower right) enzyme activity, respectively.
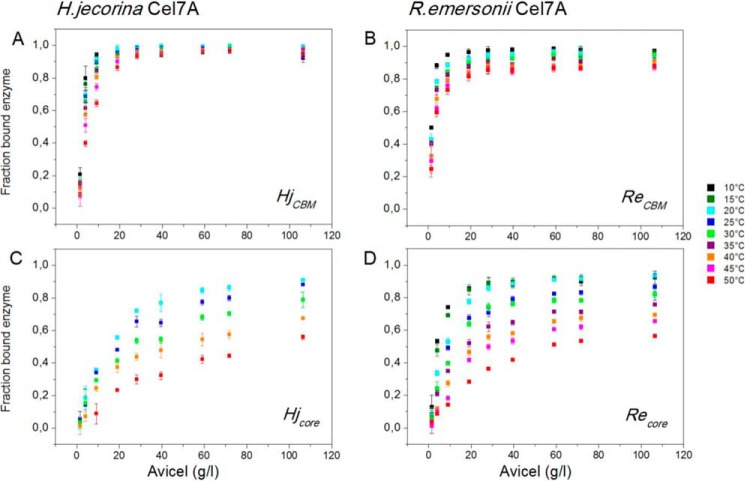
The distribution between free and adsorbed enzyme was measured in all samples at the end of the hydrolysis experiments. Results of these measurements are presented in Fig. 5, which shows the fraction of bound enzyme as a function of the substrate load. It appears that adsorption is strongly promoted by the CBM, particularly at high temperatures where a much higher fraction of the two-domain enzymes is bound compared with enzymes without a CBM. Fig. 5 also illustrates that at low substrate loads (below 30–40 g/liter) increasing temperature (at a fixed substrate load) consistently lowers the bound fraction, and this negative effect of temperature on adsorption is more pronounced for the single-domain enzymes. At higher loads of substrate where binding sites were in large excess, enzymes with a CBM were essentially fully adsorbed, and no effect of temperature could be detected. At the lower substrate loads where the effect of temperature on adsorption was largest, the CBM appeared to protect against temperature-induced affinity loss somewhat more in HjCBM compared with ReCBM. As the sequence of the linker and CBM (as well as the expression organism) were the same in both cases, this is unlikely to depend on differences in the CBM itself. It could reflect that the HjCBM enzyme with its native linker and binding module showed some kind of cooperative interaction, which was absent in the artificial fusion of the CBM-less Re wild type and the CBM from H. jecorina. To quantify the absorption data, we converted it to substrate coverage, Γ, in units of μmol of enzyme/g of cellulose using the relationship Γ = (E0 − [E])/S0 where [E] is the measured free enzyme concentration. We plotted Γ against the free enzyme concentration, [E] (not shown), and determined the slope for [E] → 0. This is the so-called partitioning coefficient, Kp, which is often used as a measure of the overall affinity of a cellulase for the cellulose surface (42, 43). Values of Kp are listed in Table 1.
FIGURE 5.
Fraction of bound Cel7A in the hydrolysis samples as function of substrate load at temperatures from 10 to 50 °C. The total enzyme concentration was 400 nm in all samples, and the measurements were made after 1-h contact time. All points are average ± S.D. (error bars) for triplicate measurements.
Discussion
The effect of temperature on cellulolytic enzyme activity is of direct interest within different research fields. For example, it has been hypothesized that temperature activation of cellulases and related glycoside hydrolases that decompose soil organic matter could shift natural carbon stocks toward more atmospheric CO2 as the climate gets warmer (44). If indeed so, this would generate a positive feedback loop for global warming, and any attempt to predict such effects relies heavily on an understanding of the activation of relevant enzymes (1). Within biotechnology, the interest in cellulases is primarily driven by their use in emerging industries that produce bioethanol from lignocellulosic feedstock. In the industrial saccharification process, it is desirable to use as high a temperature and solid loadings as possible (45, 46), and many studies have therefore investigated optimal temperatures both for wild types and thermostable variants (8–13). Although ill-defined from a rigorous point of view (47), the optimal temperature specifies the location of the maximum that usually appears when the activity against a certain substrate is plotted as a function of temperature. It occurs as a result of two independent processes. Catalytic reactions are accelerated as temperature increases, but at the optimal temperature, this is balanced out by thermal inactivation of the enzyme, and at still higher temperatures, activity is reduced due to rapid inactivation. The balance of these two processes must be considered in any enzyme temperature study. For the current enzymes, earlier work has suggested a very small degree of inactivation of HjCBM under the conditions studied here (1-h contact with Avicel and T < 50 °C) (48), and as R. emersonii Cel7A is significantly more thermostable than the H. jecorina enzyme (49), it appears safe to assume that thermal inactivation can be generally neglected in this work. Therefore, we will henceforth interpret the results with respect to the effect of temperature on catalysis and adsorption without interference from enzyme inactivation.
Cellulases perform heterogeneous catalysis, and this implies that only surface-adsorbed enzymes are potentially active. Many earlier works have shown that the adsorbed population of both two-domain Cel7A and isolated CBM decreases with increasing temperature (42, 43, 50, 52, 53), and it follows that temperature-activity relationships on insoluble substrate may be seen as a balance between accelerated reaction steps on one hand and a shift toward less surface-adsorbed (potentially active) enzyme on the other hand. The results in Table 1 allow evaluation of these two contributions. Thus, the parameter pVmax/E0 is the specific rate at saturating substrate loads where all enzyme is per definition adsorbed. We found strong thermoactivation for this parameter with Q10 values of 2.1–2.6, and we interpret this as a measure of the temperature-induced acceleration of the catalytic reaction devoid of contributions from shifts in the adsorption equilibrium. Earlier studies on soluble substrate analogs have suggested a comparable or moderately lower temperature sensitivity of Vmax for cellobiohydrolases (1, 24, 54), and hence there were no signs of pronounced effects of an insoluble substrate on temperature activation at saturating substrate loads. Comparing the two enzymes, ReCBM and ReCORE, from a thermophile organism with the analogous enzymes from the mesophile H. jecorina suggested a higher thermoactivation for the former (pVmax had Q10 values that were higher by 0.2–0.3 unit for R. emersonii). This behavior is in line with theories suggesting stronger thermoactivation (i.e. higher activation energies) of enzymes adapted to higher temperatures (23, 55), and a similar behavior has been seen in some (54) but not all (1) earlier cellulase studies using soluble substrate analogs.
At low substrate loads, we found a very different picture with a much lower degree of temperature activation. Hence, Q10 values for pη, which is the apparent second order rate constant that governs the hydrolytic rate when S ≪ pKm, was only 1.1–1.2, and this unusually low temperature sensitivity was the result of a strong growth of pKm. This parameter typically increased by an order of magnitude upon heating from 10 to 50 °C (Table 1 and Fig. 2B), and the direct meaning of this is that the substrate load required to achieve half of the maximal rate increased about 10-fold in this temperature interval. However, as pKm defined in Equation 2 is the ratio of the off- and on-rate constants, pKm may also be interpreted as a dissociation constant for the enzyme-substrate complex, and this underscores that its stability decreased noticeably upon heating. The same conclusion was reached from the independently measured Kp values (Table 1). It should be noted that Kp corresponds to a binding constant, and hence it is the reciprocal, 1/Kp, that is relevant to compare with pKm. It appears from the results in Table 1 that 1/Kp also increased strongly with temperature, and it follows that both kinetic and adsorption data confirm the same trend of a much weaker interaction as temperature increased. This agreement also offers model-independent support for the general validity of the kinetic analysis. Further inspection of Figs. 2B and 3B shows that the temperature-induced loss of affinity was particularly strong for one-domain enzymes. In fact, for enzymes without a CBM, Q10 for pη was essentially 1 (Fig. 3C), thus suggesting that their activity was almost independent of temperature at very low substrate loads. We interpret this behavior as the result of an almost complete balancing between the normal acceleration of the catalytic steps on one hand and a shift toward less enzyme-substrate complex on the other hand. If we combine the observed effects of the CBM at high and low substrate loads, it appears that the binding module exerted a dual role on temperature activation. On one hand, the CBM hampered activation because it reduced growth in the maximal rate (Q10 for pVmax was smaller for the two-domain enzymes). On the other hand, it favored activation by diminishing the tendency of desorption from the substrate as temperature increased (Q10 for pKm was also smaller for two-domain enzymes). As a consequence, the CBM favored activity at low, but not at high, substrate loads as illustrated in Fig. 4. The same conclusion has been reached previously by Viikari and co-workers (56–58), who studied a number of systems and conditions including lignocellulosic substrates and long term experiments with significant conversion. These workers concluded that the increased probability of enzymes to find the substrate at high loads would compensate for the lower affinity of one-domain enzymes (56). This argument is along the lines of Le Chatelier's principle, which stipulates a shift toward the adsorbed form at high loads, and the results in Table 1 are compatible with this. However, the current results suggest that other factors must be considered to rationalize relationships between the CBM and catalytic efficacy. Thus, at saturating substrate loads, the one-domain enzymes were consistently about 2-fold faster than enzymes with CBM, and this cannot depend on adsorption because the whole enzyme population (whether one- or two-domain) is adsorbed at saturation. We conclude that the CBM effects both adsorption and catalysis: it promotes adsorption and hence enzymatic activity at low substrate loads where “finding” the substrate is critical but exerts a negative effect on the rate of catalysis, which becomes evident at high loads. For a processive enzyme, this effect of the CBM could rely on a slow dissociation (i.e. low pkoff in Scheme 1). Several studies have suggested that at least under some conditions the overall hydrolytic rate is governed by the release of unproductively bound enzymes, which although associated with a cellulose strand are prevented from further processive movement by irregularities on the cellulose surface (39, 59–61). If CBM-cellulose interactions indeed contribute to a slower dissociation, higher maximal rates for the one-domain variants could reflect faster dissociation of unproductively bound enzyme (and hence earlier recruitment for new attacks). This interpretation parallels a recent study where the substrate affinity of HjCBM was lowered by the mutation W38A in the catalytic domain (25). Kinetic analysis of this variant at 25 °C showed a 4-fold increase in pKm and a 2-fold increase in pVmax compared with the wild type. Interestingly, these relative changes are essentially identical to those seen when comparing HjCBM and HjCORE at 25 °C (Table 1). In other words, we find the same inverse correlation between affinity and maximal rate in variants that are modified in very different ways, and although two examples are obviously not sufficient to make any general conclusions, the coincidence supports both the interpretation that pkoff is rate-limiting and the conclusion that the CBM makes a negative contribution to this rate constant (and hence slows down dissociation).
In conclusion, we have measured activity and adsorption on insoluble substrate of four related cellobiohydrolases in small increments of temperature and substrate load and used a steady state model for processive hydrolysis to analyze their temperature activation. The most temperature-sensitive parameter was the maximal rate, which showed Q10 values up to 2.6. This degree of activation is comparable with or slightly higher than results from earlier studies on cellobiohydrolases acting on soluble substrate analogs. We suggest that this provides a measure of the thermal acceleration of the reactions steps in the investigated enzymes. Other factors including temperature-induced changes in the structure of cellulose cannot be ruled out but appear unlikely. Thus, although earlier works have identified such changes, they were only seen at far higher temperatures (51). Heating was associated with a pronounced reduction in substrate affinity, and this was reflected in pKm and 1/Kp, which both grew by about an order of magnitude between 10 and 50 °C. One consequence of this was that the overall hydrolytic rate showed little and in some cases no (see Fig. 2C) increase with temperature in dilute substrate suspensions. The investigated cellobiohydrolases were chosen to elucidate whether the CBM and natural adaptation to higher temperature influenced temperature activation. We consistently found that the CBM lowered both pVmax and its increment with temperature. This negative contribution of the CBM to activity was counteracted by its promotion of substrate affinity, and as a result, the presence of a CBM was an advantage for activity at low substrate loads but a disadvantage for activity at high loads. The substrate load where the effect of the CBM on activity changed sign increased strongly with temperature (Fig. 4). The effect of the CBM was similar for the Hj and Re enzymes, although the former is naturally evolved with a CBM, whereas the latter is not. Hence, we did not find signs of additional interactions in the Re enzyme, which might have evolved to compensate for the lack of a CBM. The results revealed a moderately higher temperature activation (0.2–0.3 Q10 unit) of pVmax for thermophilic (Re) enzymes compared with the mesophilic (Hj) analogs, and this behavior was in line with general theories on temperature adaptation of enzymes and may suggest that enzymes from thermophilic organisms are beneficial for technical applications with respect to both physical stability and temperature activation.
Author Contributions
T. H. S. and P. W. conceived the study and wrote the paper. N. C. B. contributed to the analysis and interpretation of data based on the steady state model. M. S. W. designed, constructed, and expressed the WT and mutant proteins and revised the work associated with the writing of the article. S. F. B. purified the mutant proteins and designed, performed, and analyzed the experiments in Fig. 5. K. B. and T. H. S. coordinated and made the strategy for the experimental setup. All authors analyzed the results and approved the final version of the manuscript.
This work was supported by Danish Council for Strategic Research, Program Commission on Sustainable Energy and Environment Grants 2104-07-0028 and 11-116772 (to P. W.) and by Carlsberg Foundation Grant 2013-01-0208 (to P. W.). Novozymes is a major enzyme-producing company.
The Q10 values can be estimated from the activation energies, ln(Q10) = 10Ea/RT2, at a given temperature. A statistically better approach is to plot the natural logarithm of a parameter against the temperature (rather than the reciprocal temperature). The slope of this plot, α, is a direct (temperature-independent) measure of Q10, ln(Q10) = 10α. We tried both ways and found the same results probably because the investigated temperature interval is quite small on the Kelvin scale.
- CBM
- carbohydrate binding domain
- CORE
- catalytic domain
- HjCBM
- cellobiohydrolase Cel7A from H. jecorina (wild type with CORE and CBM)
- HjCORE
- H. jecorina Cel7A variant without CBM
- ReCORE
- Cel7A from R. emersonii (wild type without CBM)
- ReCBM
- chimeric enzyme with CORE from R. emersonii and CBM from H. jecorina
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. German D. P., Marcelo K. R. B., Stone M. M., Allison S. D. (2012) The Michaelis-Menten kinetics of soil extracellular enzymes in response to temperature: a cross-latitudinal study. Glob. Chang. Biol. 18, 1468–1479 [Google Scholar]
- 2. Wilson D. B. (2009) Cellulases and biofuels. Curr. Opin. Biotechnol. 20, 295–299 [DOI] [PubMed] [Google Scholar]
- 3. Horn S. J., Sikorski P., Cederkvist J. B., Vaaje-Kolstad G., Sørlie M., Synstad B., Vriend G., Vårum K. M., Eijsink V. G. (2006) Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc. Natl. Acad. Sci. U.S.A. 103, 18089–18094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou W., Irwin D. C., Escovar-Kousen J., Wilson D. B. (2004) Kinetic studies of Thermobifida fusca Cel9A active site mutant enzymes. Biochemistry 43, 9655–9663 [DOI] [PubMed] [Google Scholar]
- 5. Payne C. M., Knott B. C., Mayes H. B., Hansson H., Himmel M. E., Sandgren M., Ståhlberg J., Beckham G. T. (2015) Fungal cellulases. Chem. Rev. 115, 1308–1448 [DOI] [PubMed] [Google Scholar]
- 6. Wolfenden R., Snider M. J. (2001) The depth of chemical time and the power of enzymes as catalysts. Acc. Chem. Res. 34, 938–945 [DOI] [PubMed] [Google Scholar]
- 7. Wolfenden R., Lu X. D., Young G. (1998) Spontaneous hydrolysis of glycosides. J. Am. Chem. Soc. 120, 6814–6815 [Google Scholar]
- 8. Heinzelman P., Snow C. D., Wu I., Nguyen C., Villalobos A., Govindarajan S., Minshull J., Arnold F. H. (2009) A family of thermostable fungal cellulases created by structure-guided recombination. Proc. Natl. Acad. Sci. U.S.A. 106, 5610–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Voutilainen S. P., Puranen T., Siika-Aho M., Lappalainen A., Alapuranen M., Kallio J., Hooman S., Viikari L., Vehmaanperä J., Koivula A. (2008) Cloning, expression, and characterization of novel thermostable family 7 cellobiohydrolases. Biotechnol. Bioeng. 101, 515–528 [DOI] [PubMed] [Google Scholar]
- 10. Voutilainen S. P., Murray P. G., Tuohy M. G., Koivula A. (2010) Expression of Talaromyces emersonii cellobiohydrolase Cel7A in Saccharomyces cerevisiae and rational mutagenesis to improve its thermostability and activity. Protein Eng. Des. Sel. 23, 69–79 [DOI] [PubMed] [Google Scholar]
- 11. Wu I., Arnold F. H. (2013) Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures. Biotechnol. Bioeng. 110, 1874–1883 [DOI] [PubMed] [Google Scholar]
- 12. Day A. G., Goedegebuur F., Gualfetti P., Mitchinson C., Neefe P., Sandgren M., Shaw A., Stahlberg J. ( February 26, 2004) World Intellectual Property Organization Patent WO2004016760 [Google Scholar]
- 13. Teter S., Cherry J., Ward C., Jones A., Harris P., Yi J. ( April 2, 2009)World Intellectual Property Organization Patent WO2005030926 [Google Scholar]
- 14. Viikari L., Alapuranen M., Puranen T., Vehmaanperä J., Siika-Aho M. (2007) Thermostable enzymes in lignocellulose hydrolysis. Adv. Biochem. Eng. Biotechnol. 108, 121–145 [DOI] [PubMed] [Google Scholar]
- 15. Vásquez M. P., da Silva J. N., de Souza M. B. Jr., Pereira N. Jr. (2007) Enzymatic hydrolysis optimization to ethanol production by simultaneous saccharification and fermentation. Appl. Biochem. Biotechnol. 137, 141–153 [DOI] [PubMed] [Google Scholar]
- 16. Zhang Y., Xu J. L., Qi W., Yuan Z. H., Zhuang X. S., Liu Y., He M. C. (2012) A fractal-like kinetic equation to investigate temperature effect on cellulose hydrolysis by free and immobilized cellulase. Appl. Biochem. Biotechnol. 168, 144–153 [DOI] [PubMed] [Google Scholar]
- 17. Hu G., Heitmann J. A. Jr., Rojas O. J. (2009) In situ monitoring of cellulase activity by microgravimetry with a quartz crystal microbalance. J. Phys. Chem. B 113, 14761–14768 [DOI] [PubMed] [Google Scholar]
- 18. Drissen R. E. T., Maas R. H. W., Van Der Maarel M., Kabel M. A., Schols H. A., Tramper J., Beeftink H. H. (2007) A generic model for glucose production from various cellulose sources by a commercial cellulase complex. Biocatal. Biotransform. 25, 419–429 [Google Scholar]
- 19. Zhang S., Wolfgang D. E., Wilson D. B. (1999) Substrate heterogeneity causes the nonlinear kinetics of insoluble cellulose hydrolysis. Biotechnol. Bioeng. 66, 35–41 [DOI] [PubMed] [Google Scholar]
- 20. Subhedar P. B., Gogate P. R. (2014) Enhancing the activity of cellulase enzyme using ultrasonic irradiations. J. Mol. Catal. B Enzym. 101, 108–114 [Google Scholar]
- 21. Deng S. P., Tabatabai M. A. (1994) Cellulase activity of soils. Soil Biol. Biochem. 26, 1347–1354 [Google Scholar]
- 22. Ng T. K., Zeikus J. G. (1981) Comparison of extracellular cellulase activities of Clostridium thermocellum LQRI and Trichoderma reesei QM9414. Appl. Environ Microbiol. 42, 231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lonhienne T., Gerday C., Feller G. (2000) Psychrophilic enzymes: revisiting the thermodynamic parameters of activation may explain local flexibility. Biochim. Biophys. Acta 1543, 1–10 [DOI] [PubMed] [Google Scholar]
- 24. Stone M. M., Weiss M. S., Goodale C. L., Adams M. B., Fernandez I. J., German D. P., Allison S. D. (2012) Temperature sensitivity of soil enzyme kinetics under N-fertilization in two temperate forests. Glob. Chang. Biol. 18, 1173–1184 [Google Scholar]
- 25. Kari J., Olsen J., Borch K., Cruys-Bagger N., Jensen K., Westh P. (2014) Kinetics of cellobiohydrolase (Cel7A) variants with lowered substrate affinity. J. Biol. Chem. 289, 32459–32468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ye Z., Berson R. E. (2014) Factors affecting cellulose hydrolysis based on inactivation of adsorbed enzymes. Bioresour. Technol. 167, 582–586 [DOI] [PubMed] [Google Scholar]
- 27. Brown R. F., Agbogbo F. K., Holtzapple M. T. (2010) Comparison of mechanistic models in the initial rate enzymatic hydrolysis of AFEX-treated wheat straw. Biotechnol. Biofuels 3, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cruys-Bagger N., Elmerdahl J., Praestgaard E., Borch K., Westh P. (2013) A steady-state theory for processive cellulases. FEBS J. 280, 3952–3961 [DOI] [PubMed] [Google Scholar]
- 29. Houbraken J., Spierenburg H., Frisvad J. C. (2012) Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie Van Leeuwenhoek 101, 403–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grassick A., Murray P. G., Thompson R., Collins C. M., Byrnes L., Birrane G., Higgins T. M., Tuohy M. G. (2004) Three-dimensional structure of a thermostable native cellobiohydrolase, CBH IB, and molecular characterization of the cel7 gene from the filamentous fungus, Talaromyces emersonii. Eur. J. Biochem. 271, 4495–4506 [DOI] [PubMed] [Google Scholar]
- 31. Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., Claeyssens M. (1988) Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur. J. Biochem. 170, 575–581 [DOI] [PubMed] [Google Scholar]
- 32. Divne C., Ståhlberg J., Reinikainen T., Ruohonen L., Pettersson G., Knowles J. K., Teeri T. T., Jones T. A. (1994) The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science 265, 524–528 [DOI] [PubMed] [Google Scholar]
- 33. Borch K., Jensen K., Krogh K., McBrayer B., Westh P., Kari J., Olsen J. P., Sorensen T. H., Windahl M. S., Xu H. ( September 12, 2014) World Intellectual Property Organization Patent WO2014138672 [Google Scholar]
- 34. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J. M., ed) pp. 571–607, Humana Press, Totowa, NJ [Google Scholar]
- 35. Lever M. (1972) A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 47, 273–279 [DOI] [PubMed] [Google Scholar]
- 36. Murphy L., Bohlin C., Baumann M. J., Olsen S. N., Sørensen T. H., Anderson L., Borch K., Westh P. (2013) Product inhibition of five Hypocrea jecorina cellulases. Enzyme Microb. Technol. 52, 163–169 [DOI] [PubMed] [Google Scholar]
- 37. Teugjas H., Väljamäe P. (2013) Product inhibition of cellulases studied with C-14-labeled cellulose substrates. Biotechnol. Biofuels 6, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gruno M., Väljamäe P., Pettersson G., Johansson G. (2004) Inhibition of the Trichoderma reesei cellulases by cellobiose is strongly dependent on the nature of the substrate. Biotechnol. Bioeng. 86, 503–511 [DOI] [PubMed] [Google Scholar]
- 39. Praestgaard E., Elmerdahl J., Murphy L., Nymand S., McFarland K. C., Borch K., Westh P. (2011) A kinetic model for the burst phase of processive cellulases. FEBS J. 278, 1547–1560 [DOI] [PubMed] [Google Scholar]
- 40. Horn S. J., Sørlie M., Vårum K. M., Väljamäe P., Eijsink V. G. (2012) Measuring processivity. Methods Enzymol. 510, 69–95 [DOI] [PubMed] [Google Scholar]
- 41. Fersht A. (1998) Structure and Mechanism in Protein Science: a Guide to Enzyme Catalysis and Protein Folding, W. H. Freeman, New York [Google Scholar]
- 42. Linder M., Teeri T. T. (1996) The cellulose-binding domain of the major cellobiohydrolase of Trichoderma reesei exhibits true reversibility and a high exchange rate on crystalline cellulose. Proc. Natl. Acad. Sci. U.S.A. 93, 12251–12255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Palonen H., Tenkanen M., Linder M. (1999) Dynamic interaction of Trichoderma reesei cellobiohydrolases Cel6A and Cel7A and cellulose at equilibrium and during hydrolysis. Appl. Environ. Microbiol. 65, 5229–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davidson E. A., Janssens I. A. (2006) Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440, 165–173 [DOI] [PubMed] [Google Scholar]
- 45. Larsen J., Petersen M. O., Thirup L., Li H. W., Iversen F. K. (2008) The IBUS process—lignocellulosic bioethanol close to a commercial reality. Chem. Eng. Technol. 31, 765–772 [Google Scholar]
- 46. Jorgensen H., Kristensen J. B., Felby C. (2007) Enzymatic conversion of lignocellulose into fermentable sugars: challenges and opportunities. Biofuel. Bioprod. Bioref. 1, 119–134 [Google Scholar]
- 47. Laidler K. J., Peterman B. F. (1979) Temperature effects in enzyme kinetics. Methods Enzymol. 63, 234–257 [DOI] [PubMed] [Google Scholar]
- 48. Alasepp K., Borch K., Cruys-Bagger N., Badino S., Jensen K., Sørensen T. H., Windahl M. S., Westh P. (2014) In situ stability of substrate-associated cellulases studied by DSC. Langmuir 30, 7134–7142 [DOI] [PubMed] [Google Scholar]
- 49. Voutilainen S. P., Nurmi-Rantala S., Penttilä M., Koivula A. (2014) Engineering chimeric thermostable GH7 cellobiohydrolases in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 98, 2991–3001 [DOI] [PubMed] [Google Scholar]
- 50. Moran-Mirabal J. M., Bolewski J. C., Walker L. P. (2011) Reversibility and binding kinetics of Thermobifida fusca cellulases studied through fluorescence recovery after photobleaching microscopy. Biophys. Chem. 155, 20–28 [DOI] [PubMed] [Google Scholar]
- 51. Watanabe A., Morita S., Ozaki Y. (2006) Temperature-dependent structural changes in hydrogen bonds in microcrystalline cellulose studied by infrared and near-infrared spectroscopy with perturbation-correlation moving-window two-dimensional correlation analysis. Appl. Spectrosc. 60, 611–618 [DOI] [PubMed] [Google Scholar]
- 52. Ooshima H., Sakata M., Harano Y. (1983) Adsorption of cellulase from Trichoderma viride on cellulose. Biotechnol. Bioeng. 25, 3103–3114 [DOI] [PubMed] [Google Scholar]
- 53. Colussi F., Sørensen T. H., Alasepp K., Kari J., Cruys-Bagger N., Windahl M. S., Olsen J. P., Borch K., Westh P. (2015) Probing substrate interactions in the active tunnel of a catalytically deficient cellobiohydrolase (Cel7). J. Biol. Chem. 290, 2444–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garsoux G., Lamotte J., Gerday C., Feller G. (2004) Kinetic and structural optimization to catalysis at low temperatures in a psychrophilic cellulase from the Antarctic bacterium Pseudoalteromonas haloplanktis. Biochem. J. 384, 247–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Somero G. N. (2004) Adaptation of enzymes to temperature: searching for basic “strategies.” Comp. Biochem. Physiol. B Biochem. Mol. Biol. 139, 321–333 [DOI] [PubMed] [Google Scholar]
- 56. Le Costaouëc T., Pakarinen A., Várnai A., Puranen T., Viikari L. (2013) The role of carbohydrate binding module (CBM) at high substrate consistency: comparison of Trichoderma reesei and Thermoascus aurantiacus Cel7A (CBHI) and Cel5A (EGII). Bioresour. Technol. 143, 196–203 [DOI] [PubMed] [Google Scholar]
- 57. Pakarinen A., Haven M. O., Djajadi D. T., Várnai A., Puranen T., Viikari L. (2014) Cellulases without carbohydrate-binding modules in high consistency ethanol production process. Biotechnol. Biofuels 7, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Várnai A., Siika-Aho M., Viikari L. (2013) Carbohydrate-binding modules (CBMs) revisited: reduced amount of water counterbalances the need for CBMs. Biotechnol. Biofuels 6, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cruys-Bagger N., Elmerdahl J., Praestgaard E., Tatsumi H., Spodsberg N., Borch K., Westh P. (2012) Pre-steady-state kinetics for hydrolysis of insoluble cellulose by cellobiohydrolase Cel7A. J. Biol. Chem. 287, 18451–18458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jalak J., Väljamäe P. (2010) Mechanism of initial rapid rate retardation in cellobiohydrolase catalyzed cellulose hydrolysis. Biotechnol. Bioeng. 106, 871–883 [DOI] [PubMed] [Google Scholar]
- 61. Eriksson T., Karlsson J., Tjerneld F. (2002) A model explaining declining rate in hydrolysis of lignocellulose substrates with cellobiohydrolase I (cel7A) and endoglucanase I (cel7B) of Trichoderma reesei. Appl. Biochem. Biotechnol. 101, 41–60 [DOI] [PubMed] [Google Scholar]
- 62. Sørensen T. H., Cruys-Bagger N., Borch K., Westh P. (2015) Free energy diagram for the heterogeneous enzymatic hydrolysis of glycosidic bonds in cellulose. J. Biol. Chem. 290, 22203–22211 [DOI] [PMC free article] [PubMed] [Google Scholar]