Figure 1.
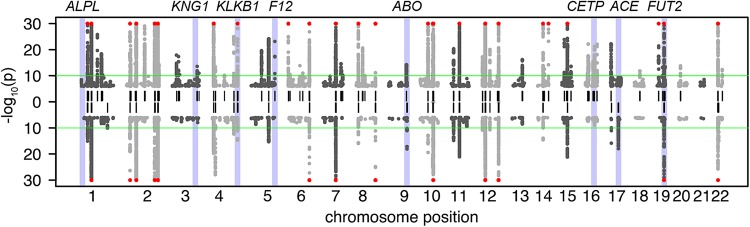
Manhattan plot of the mGWAS by Shin et al. (23). Upward pointing P-values: TwinsUK cohort, downward pointing P-values: KORA population study. Only SNPs with association to raw metabolites (P < 10−6) are displayed (no ratios). The green line indicates the genome-wide significance cutoff for the P-value (P < 10−10). Loci that reach genome-wide significance in either cohort are indicated by a short vertical black line. Loci with P-values <10−30 are indicated with a red symbol. Loci that are further discussed in Figure 3 are highlighted and annotated [figure adapted from the Supplementary Material, Fig. S2 by Shin et al., Nature Genetics, 2014 (23)].
