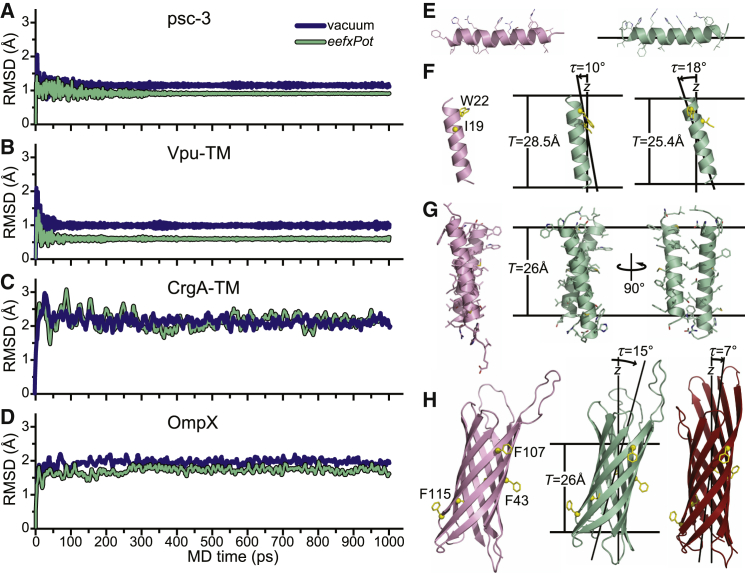
Figure 2.
MD simulations of the experimentally determined structures of psc-3, Vpu-TM, CrgA-TM, and OmpX, performed with eefxPot or vacuum in the absence of experimental restrains. Simulations were performed at 300 K in Cartesian space. Membranes are depicted as horizontal lines separated by thickness T. (A–D) Plots of structural accuracy as a function of MD time with eefxPot (green) or vacuum (blue). Accuracy is reported as the backbone atom (N, CA, and C) RMSD to the experimental structure deposited in the PDB. (E) Structure of psc-3 in DMPC/DMPG-oriented bilayers (48) taken directly from the PDB (2MCW, pink) or after eefxPot simulation in a 25.4 Å membrane (green). (F) Structure of Vpu-TM in DOPC-oriented bilayers (49) taken directly from the PDB (1PI7, pink) or after eefxPot simulation in 28.5 Å or 25.4 Å membranes (green). (G) Structure of CrgA-TM in DOPC-oriented bilayers (50) taken directly from the PDB (2MMU, pink) or after eefxPot simulation in a 26 Å membrane (green). (H) Structure of OmpX in DMPC/DMPG nanodiscs (51) taken directly from the PDB (2M06, pink) or after eefxPot simulation in a 26 Å membrane (green). The crystal structure (57) (1QJ8, red) is shown in the membrane position derived by rigid-body orientation with solid-state NMR restraints (56). To see this figure in color, go online.