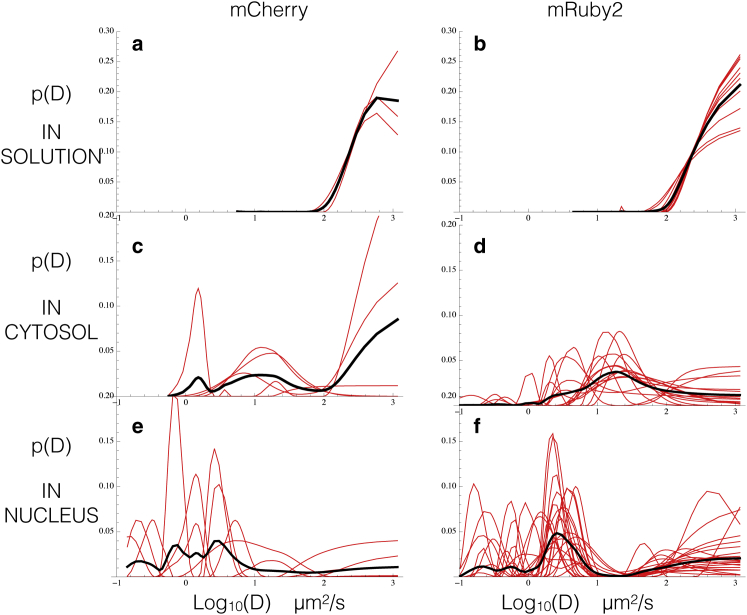
Figure 6.
We extract probability distributions of diffusion coefficients from FCS curves. To extract p(D) s, we used FCS data acquired for mCherry and mRuby2 diffusing freely in solution (a and b), and for BZip protein domains tagged with mCherry and mRuby2 diffusing, whereas in the cytosol (c and d) and in the nucleus but far from regions of heterochromatin (e and f) 56. The p(D) s for the freely diffusing FPs in solution exhibit superdiffusive plateaus due to flickering and triplet corrections, with the actual free diffusion coefficient appearing as a lower bound. In the cytosolic and nuclear data, these plateaus are still present for the FP-tagged BZips but they are accompanied by rich landscapes that capture the many different interactions between the FP-tagged BZips and their environment detailed in the main body. Red curves represent the p(D) s extracted from individual data sets; Black curves are the average of all underlying red curves. The total number of data sets averaged is per plot: (a) 3, (b) 9, (c) 5, (d) 16, (e) 7, and (f) 21. In cases where only a few data sets were available the average occasionally exhibits features that are only found in one data set; we consider such features to be most likely artifacts of heterogeneity between different ROIs in the cytosol or nucleus and ignore them in our analysis. In cases where more data sets are averaged such single data set features are naturally muted. We note that, predictably, in solution the p(D) s are far less noisy as compared to the heterogeneous environment of a living cell. To see this figure in color, go online.