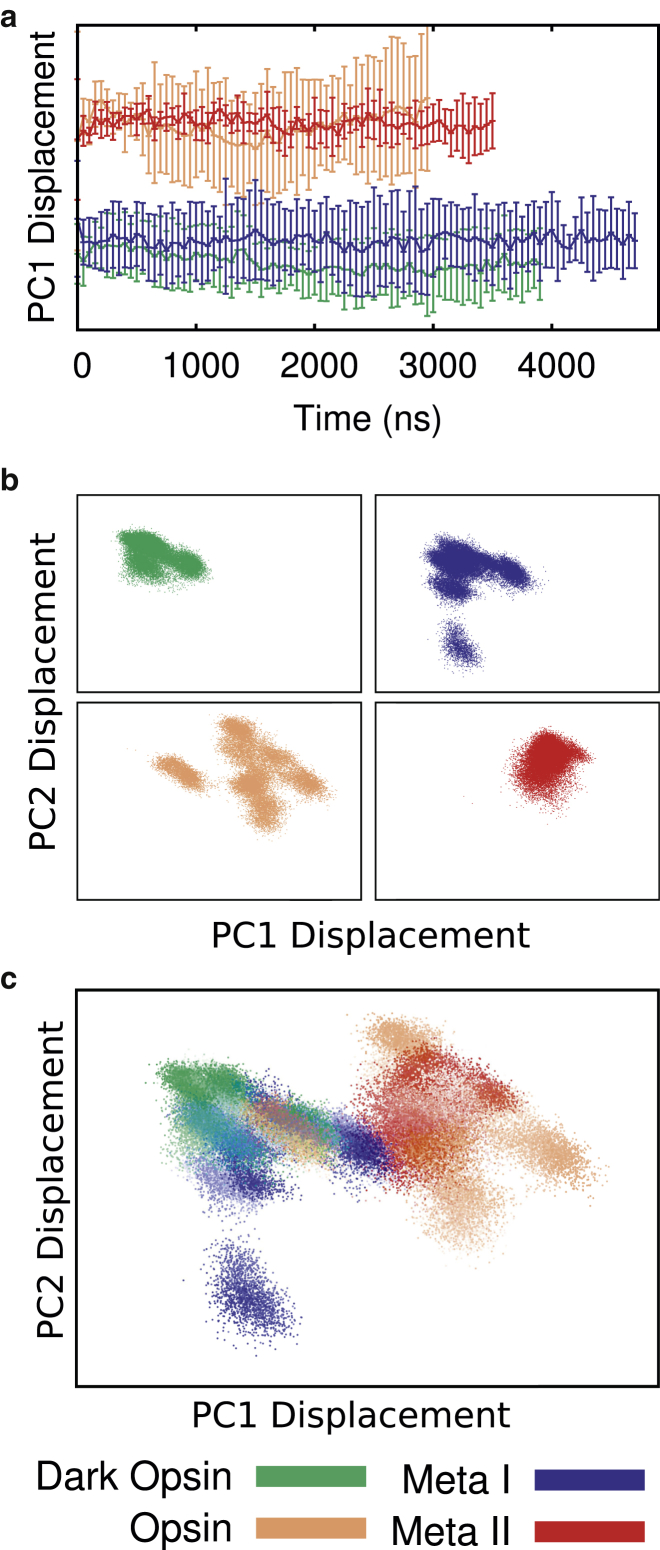
Figure 5.
Ensemble-based principal component analysis was conducted on the transmembrane α-carbons of all 24 trajectories in aggregate, so the results depict how each simulation is projected onto the same basis set. Each of the trajectories used in this analysis is labeled according to its ensemble as shown in the key at the bottom of the figure. (a) Average projection of trajectory time course onto principal component one. Simulations are grouped by the four starting structures outlined in Table 1. Curves show the average across all six trajectories for each ensemble, with error bars showing the average of the standard deviations in each dataset, to emphasize which trajectories are more heterogeneous. (b and c) Projection onto the first two principal components. Each data point represents a structure from the ensemble of trajectories (sampled at 1 ns a piece) and colored according to ensemble. The x axis represents displacement along PC1, the y axis displacement along PC2. (b) Each ensemble is shown separately on the same scale. (c) Merged view of the result from (b). Opacity of the points shows the time evolution of each independent trajectory. To see this figure in color, go online.