Figure 1.
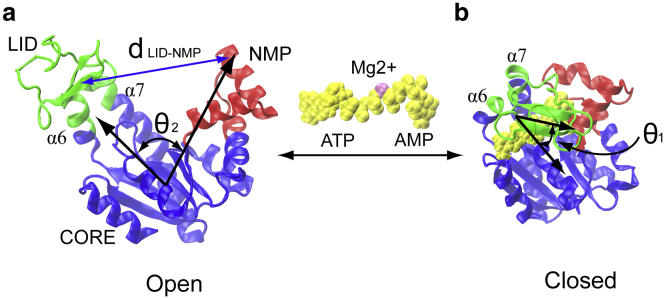
Conformational transitions in AdK: (a) the open state (e.g., PDB code: 4AKE); and (b) the closed state with ligands (e.g., PDB code: 1AKE). The enzyme consists of three well-defined domains: the rigid CORE (blue, residues 1–29, 60–121, and 160–214); nucleotide triphosphate binding domain LID (green, residues 122–159); and nucleotide monophosphate binding domain NMP (red, residues 30–59). The alpha helices α6 and α7 correspond to residues 112 to 122 and residues 160 to 189, respectively. The ligands Mg2+•ATP/AMP are represented by yellow van der Waals (VDW) spheres. The angle LID-CORE is formed by the centers of mass of the backbone of residues of LID (a.a. 123–155 (LID), hinge (a.a. 161–165), and CORE (a.a. 1–8, 79–85, 104–110, and 190–198), whereas the angle NMP-CORE is formed by the centers of mass of the backbone of residues of NMP (a.a. 50–59), CORE (a.a. 1–8, 79–85, 104–110, and 190–198), and hinge (a.a. 161–165). The variable is used to monitor the distance by the centers of mass between domains LID and NMP, respectively. To see this figure in color, go online.
