Fig. 4.
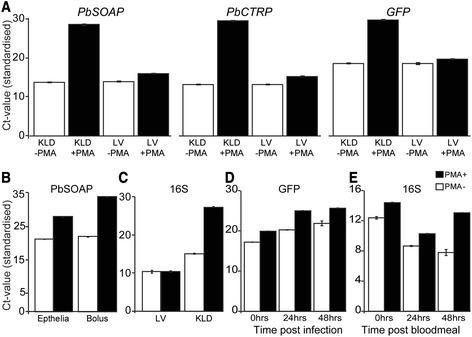
PMA-qrtPCR investigation of the inhibition of DNA amplification in killed ookinetes or bacteria during PCR. a Average cycle threshold (ct-value) for PbSOAP PbCTRP and PbGFPCON genes in heat inactivated (KLD) or live (LV) ookinetes from overnight gametocyte culture that were pre-treated with PMA. Inactivated and live ookinetes without PMA were used as a control. Samples, before DNA extraction, were spiked with an equal volume of Anopheles coluzzii homogenate with the aim to use the mosquito S7 gene as a heterologous internal standard. b Average ct-values for the PbGFP gene in the midgut contents or gut epithelium tissue from mossquitoes 24 h pi with P. berghei parasite. c Average ct-values for bacterial 16S rRNA gene from heat inactivated samples and live bacteria cultured from mosquito midgut contents (24 h after bloodmeal). Samples without PMA treatment served as a control. d Ct-values for PbGFP genes from gDNA samples that were extracted from mosquitoes that obtaned P. berghei ookinetes in their bloodmeal. e Ct-values for 16S rRNA genes in gDNA extracted from mosquitoes that obstained naïve bloodmeal. An. gambiae S7 gene served as a heterologous internal standard for all the PMA-qrtPCR reactions that were performed using samples that were isolated from mosquito tissues. Error bars represent standard deviation from at least two independent assays. Each qrPCR experiment repeated three times
