Abstract
Tumor suppressor protein p53 is a master transcription regulator, indispensable for controlling several cellular pathways. Earlier work in our laboratory led to the identification of dual internal ribosome entry site (IRES) structure of p53 mRNA that regulates translation of full-length p53 and Δ40p53. IRES-mediated translation of both isoforms is enhanced under different stress conditions that induce DNA damage, ionizing radiation and endoplasmic reticulum stress, oncogene-induced senescence and cancer. In this study, we addressed nutrient-mediated translational regulation of p53 mRNA using glucose depletion. In cell lines, this nutrient-depletion stress relatively induced p53 IRES activities from bicistronic reporter constructs with concomitant increase in levels of p53 isoforms. Surprisingly, we found scaffold/matrix attachment region-binding protein 1 (SMAR1), a predominantly nuclear protein is abundant in the cytoplasm under glucose deprivation. Importantly under these conditions polypyrimidine-tract-binding protein, an established p53 ITAF did not show nuclear-cytoplasmic relocalization highlighting the novelty of SMAR1-mediated control in stress. In vivo studies in mice revealed starvation-induced increase in SMAR1, p53 and Δ40p53 levels that was reversible on dietary replenishment. SMAR1 associated with p53 IRES sequences ex vivo, with an increase in interaction on glucose starvation. RNAi-mediated-transient SMAR1 knockdown decreased p53 IRES activities in normal conditions and under glucose deprivation, this being reflected in changes in mRNAs in the p53 and Δ40p53 target genes involved in cell-cycle arrest, metabolism and apoptosis such as p21, TIGAR and Bax. This study provides a new physiological insight into the regulation of this critical tumor suppressor in nutrient starvation, also suggesting important functions of the p53 isoforms in these conditions as evident from the downstream transcriptional target activation.
p53 is a master transcription factor and tumor suppressor. Apart from post-translational modifications of p53 and concomitant protein-protein interactions of p53 with diverse factors, translational control of p53 mRNA also plays an important role under stress conditions.1
p53 and its N-terminally truncated isoform Δ40p53 (also known as ΔN-p53 or p53/47) are translated by internal ribosome entry site (IRES)-mediated translation initiation from the same mRNA under different stress conditions that induce DNA damage, ionizing radiation and endoplasmic reticulum (ER) stress, oncogene-induced senescence and cancer.2, 3, 4, 5, 6, 7 Thus, p53 mRNA has a dual IRES structure.8 For their function, these IRESs rely on IRES trans-acting factors (ITAFs). Polypyrimidine-tract-binding protein (PTB) was shown to have differential affinity for the two IRESs of p53 and regulated p53 IRES functions by translocating from nucleus to cytoplasm on doxorubicin-induced DNA damage.3 This PTB-mediated control of p53 IRESs has been recently found to play a role in the p53-fibrillarin-rRNA methylation network.9, 10 Single-nucleotide polymorphisms in p53 5'UTR11 or in the coding region of p53 IRES7, 11, 12 decreased p53 IRES activity by altering PTB, MDM2 or hnRNPC1/C2 interactions with this regulatory region in p53 mRNA. Annexin A2 and PTB-associated splicing factor (PSF) proteins, putative p53 ITAFs, interact with p53 IRESs ex vivo in a stress-induced manner, showing greater association with the IRESs on thapsigargin treatment.13 An eIF4G homolog, death-associated protein 5 (DAP5) was demonstrated to bind to p53 IRESs and regulate the second IRES-mediated expression of Δ40p53, whereas such regulation by DAP5 of the first IRES-mediated expression of p53 was more subtle.14 hnRNPQ was demonstrated to bind to p53 5'UTR and control its translation efficiency.15 Apart from various ITAFs, p53 5'UTR is also known to bind several proteins such as RPL26,16 nucleolin,17 PDCD418 and RNPC1.19
Nutrient-limitation or starvation is also known to induce cellular stress. In Drosophila under poor nutritional conditions, FOXO (a Forkhead-box transcription factor) mediates accumulation of INR via IRES-mediated translation of the INR mRNA.20 Nutritional control of transcription/ translation via modulation of IRES activity is also exemplified by the cellular response to limited amino acid availability.21, 22 Amino acid depletion induces GCN2 kinase-mediated phosphorylation of eIF2α, leading to a global decrease in protein synthesis and induction of an adaptive survival program.21, 23 Under this condition, IRES-mediated translation of the Cat-1 and SNAT2 mRNAs occur,24, 25 thus preparing cells to transport amino acids once they become available. Methionine synthase, a key enzyme that clears intracellular homocysteine, is induced by its cofactor, vitamin B12, at a translational level through an IRES in the 5'UTR of the mRNA.26 In response to glucose deprivation, haploid Saccharomyces cerevisiae cells dramatically downregulate translation of most cellular messages,27, 28 but several yeast genes required for invasive growth, a developmental pathway induced by nutrient limitation, contain potent IRESs.29 Serum starvation of mammalian cell cultures showed induction of Bcl-2 IRES30 and activated translation of XIAP mRNA.31 IRES-mediated translation of p27Kip1 mRNA contributes to maintenance of G1 phase of the cell cycle and the expression of p27Kip1 was found to be iron sensitive.32, 33
These studies reveal a novel aspect of activation of IRES-mediated translation of eukaryotic mRNAs due to nutrient shortage, resulting in the synthesis of proteins essential for cell survival or apoptosis. Thus, it is important to investigate IRES activity of p53 mRNA in nutrient-deprived conditions. In the current study, results suggest that glucose depletion relatively induces p53 IRES activity as seen in bicistronic reporter assays. There are reports that have implicated p53 protein in binding its own RNA.34 The E3-ubiquitin ligase MDM2 is a well-known target of p53, forming a feedback loop and regulating p53 degradation. Interestingly, MDM2 has also been shown to interact with coding sequence of the IRES in p53 mRNA.12, 35, 36 A recent work suggested stress-dependent formation of a ternary complex of three proteins: p53, MDM2 and SMAR1,37 another transcriptional target of p53 that can modulate p53 transactivation potential.37, 38 We now find that SMAR1, a predominantly nuclear protein becomes abundant in the cytoplasm under glucose deprivation. Thus glucose deprivation, a form of nutrient-depletion stress, can induce p53 IRESs and also increases cytoplasmic abundance of SMAR1 that in turn binds to p53 IRESs, indicating the role of SMAR1 in controlling translation of p53 isoforms. Also, this increase in p53 isoforms is reversible suggesting that transient glucose or dietary deprivation can impinge reversibly on p53 signaling as suggested by p53-target transactivation.
Results
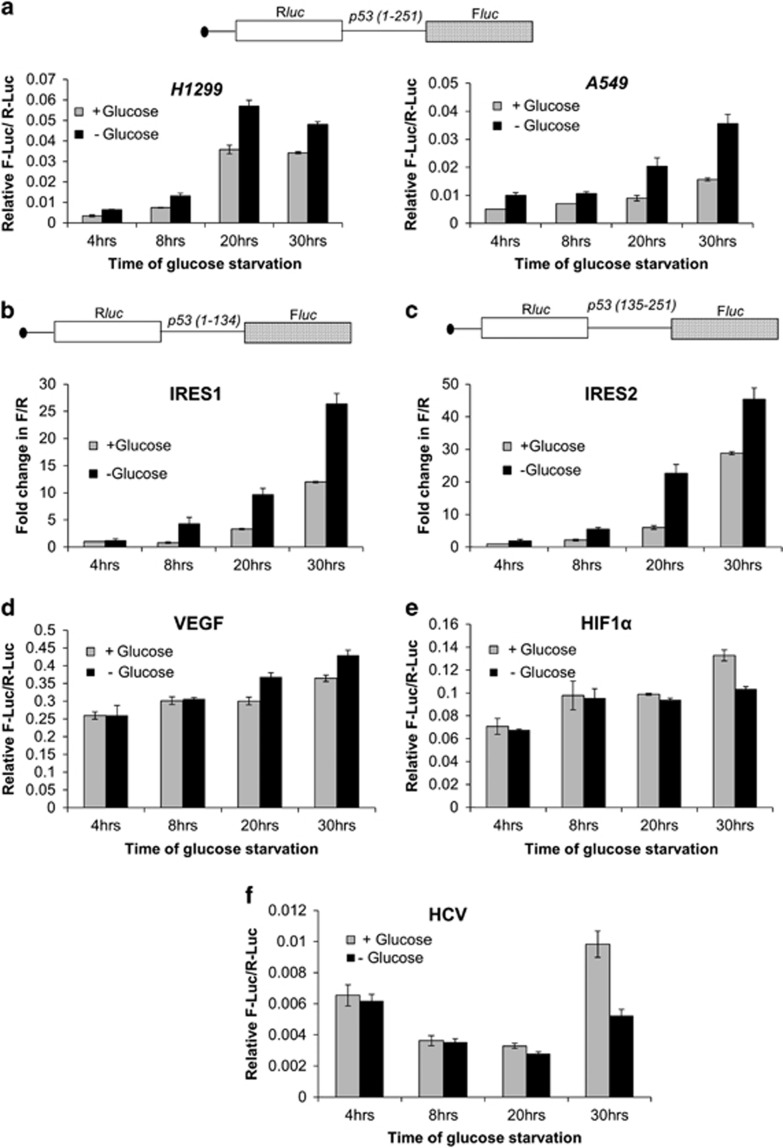
Glucose deprivation increases p53 IRES activity
p53-null H1299 cells were transfected with luciferase bicistronic constructs containing p53 1–251 RNA in the intercistronic region.7, 8, 11 Control cells and glucose-starved cells were harvested 4, 8, 20 and 30 h post-transfection. There was a consistent increase in the relative IRES activity at all these time points following glucose deprivation, with a 1.7-fold increase in activity by 20 h (Figure 1a, H1299 panel and Supplementary Table S1). Similar experiments were performed on A549 cells (with endogenous p53) and results corroborated the observation in H1299 cells. The relative IRES activity increased by 2.3-fold by 20 h and elevated IRES activity was maintained at 30 h following glucose deprivation in A549 cells (Figure 1a, A549 panel and Supplementary Table S2). In both H1299 and A549 cells, the relative increase in IRES activity also reflects a concomitant decrease in cap-dependent translation of Renilla luciferase, this being more evident in the p53-positive A549 cells, as glucose deprivation induces eIF2α phosphorylation.39 As evident from these readings, fold decrease in cap-dependent translation (R-Luc) is much higher compared to fold decrease in cap-independent translation (F-Luc), that can be interpreted as a situation wherein the IRES function maintains the translation of F-Luc. To address the relative effect of glucose deprivation on p53 IRES1 and IRES2, bicistronic plasmids containing the first (1–134) or second (135–251) IRES RNA module of p53 were transfected into H1299 cells. Similar experimental regimen as above showed that at 30 h of glucose deprivation, the activity of first IRES region increased 2.4-fold (Figure 1b and Supplementary Table S3) whereas that of second IRES module increased 1.7-fold (Figure 1c and Supplementary Table S3), a consistent increase at all the time points being a common feature of both IRESs. The response to glucose deprivation was IRES specific, relative activity of VEGF IRES (Figure 1d and Supplementary Table S4), but not HIF1α or HCV IRES (Figures 1e and f and Supplementary Tables S5 and S6, respectively), was higher in these conditions.
Figure 1.
Glucose deprivation induces p53 IRES activity. (a) Schematic representation of the bicistronic construct pRp53(1–251)F that contains 1–251 IRES RNA. Relative IRES activity is represented by the Fluc/Rluc ratio at various time points for H1299 and A549 cells, n=6. (b) Schematic representation of the bicistronic construct pRp53(1–134)F that contains the first IRES of p53, corresponding to the 134nt in the 5'UTR. Fold change in Fluc/Rluc ratio as measure of IRES activity, from fourth hour unstarved cells set at unit, at various time points in H1299 cells, n=6. (c) Schematic representation of the bicistronic construct pRp53(135–251)F that contains the second IRES of p53, corresponding to the first 117nt of the coding sequence. Fold change in Fluc/Rluc ratio as measure of IRES activity, from fourth hour unstarved cells set at unit, at various time points in H1299 cells, n=6. (d–f) Relative VEGF (d), HIF1α (e) and HCV (f) IRES activity is represented by the Fluc/Rluc ratio at various time points for H1299 cells, n=6 for each. (g) Western blot from H1299-cell extracts transfected with 14A plasmid that expresses both p53 and Δ40p53 by IRES-mediated translation, followed by glucose deprivation for 8 and 30 h. Graphs show p53 and Δ40p53 levels normalized to actin. (h) Western blot from H1299-cell extracts transfected with 14A plasmid that expresses both p53 and Δ40p53 by IRES-mediated translation, followed by glucose deprivation for 30 h and 50 μg/ml cycloheximide (CHX) treatment for next 0, 60 and 120 min. Graphs show lane-wise p53 and Δ40p53 levels normalized to actin. (i) Luciferase reporter assay for p21-promoter transactivation from H1299 cells after 24 h of pGFP-hp-p53-5'UTR-cDNA plasmid transfection and 8 h of glucose deprivation, n=6. Schematic representation of p21(WAF1) promoter-firefly luciferase and CMV-Renilla luciferase, latter acting as transfection control, are shown. Western blot in inset shows induction of p53 and Δ40p53 levels after 8 h of glucose deprivation from same lysates. (j) Western blot of H1299-cell extracts transfected with 14A-M1 or 14A-M2 plasmid (see Results section for description). Cells were starved of glucose for 8 and 30 h
For studying the effect of glucose deprivation on p53 IRES-mediated translation, pGFP-hp-p53-5'UTR-cDNA (14A) plasmid that expresses both p53 and Δ40p53 by IRES-mediated translation was transfected in H1299 cells followed by glucose deprivation for 8 h and 30 h. There was discernible increase in the steady-state levels of both the isoforms in these time points (Figure 1g). To investigate whether such an increase was due to greater stability of these isoforms after glucose deprivation, time dependent of cycloheximide (CHX, translation-initiation blocker) treatment was done 30 h post-glucose deprivation. Interestingly, the induction in the steady-state levels of p53 and Δ40p53 (Figure 1h, lane 1 versus 4) was completely abrogated post-CHX treatment (Figure 1h, lanes 5 and 6) suggesting that such induction is possibly at the level of translation initiation, which is IRES mediated in this experimental setting. Downstream to glucose-deprivation-mediated p53 induction, p21 promoter was found to be significantly activated in a luciferase reporter assay (Figure 1i).
The first transactivation domain (TAD-I) of p53 protein is the docking site for MDM2-E3 ligase that ubiquitinylates p53 and targets it for degradation.40 It was thus imperative to delineate if inhibition of MDM2 function on glucose deprivation results in elevated p53 stability. Two constructs as shown in Figure 1j were used, one would produce only p53 as second ATG was mutated (pGFP-hp-p53-5'UTR-(A252G/T253C/G254T)-cDNA or 14A-M1 plasmid), while the other would produce only Δ40p53 as the first ATG was mutated (pGFP-hp-p53-5'UTR(A135T)-cDNA or 14A-M2 plasmid), both plasmids would employ entire (1–251) IRES to direct synthesis of the protein. When cells were deprived of glucose for 8 or 30 h, there was increase in the levels p53 and also of Δ40p53 (Figure 1j). Since, Δ40p53 isoform lacks TAD-I and cannot bind MDM2, increase in its levels occurs apparently rules out possible physical participation of MDM2 in elevating steady-state levels of p53 isoforms on glucose deprivation. This also suggests that apart from any increase in protein stability that might be attributed to glucose-starvation-induced full-length p53 levels, translation of p53 mRNA (which is IRES mediated in this experiment) is also essential for such induction.
SMAR1 knockdown inhibits p53 IRES function
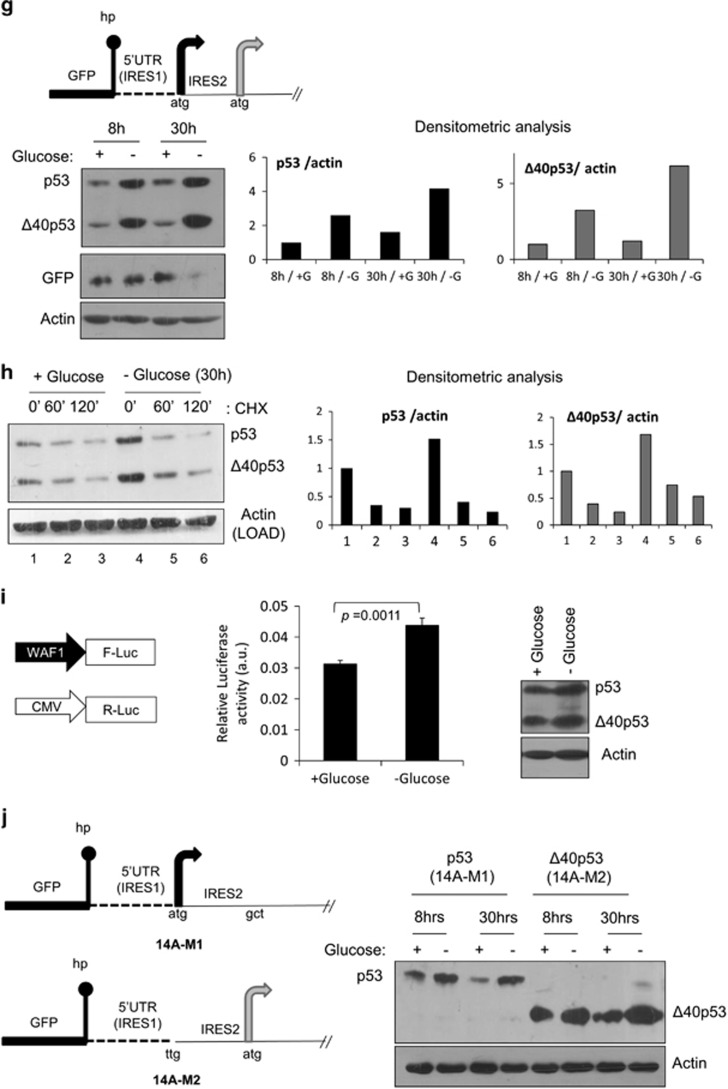
In order to determine whether SMAR1 participates in IRES-mediated translation of p53 mRNA in a more direct way, the effect of SMAR1 knockdown on the IRES-mediated expression of p53 isoforms from pGFP-hp-p53-5'UTR cDNA was assessed. An siRNA-mediated-dose-dependent decrease was seen in the levels of p53 and Δ40p53 (Figure 2a) but not in cap-dependent β-galactosidase expression (Figure 2a inset), suggesting that knocking down SMAR1 specifically affected the IRES-dependent synthesis of p53 isoforms. Similar siRNA-mediated knockdown experiments with pRp53(1–251)F bicistronic construct, representing both modules of p53 IRES is needed for complete IRES functionality in Δ40p53 synthesis,3, 8 decreased IRES activity by nearly 30% (Figure 2b and Supplementary Table S7) and SMAR1 knockdown was checked by immunoblotting (Figure 2b, inset). HCT116-p53+/+ colon carcinoma cells (like A549 non-small-cell lung carcinoma cells used) also have endogenous levels of p53 and Δ40p53, in which expression of the shorter isoform is completely IRES dependent. siRNA-mediated knockdown of SMAR1 could downregulate Δ40p53 levels in these cells (Supplementary Figure S8A). In these cells also, there was a dose-dependent decrease in IRES activity from pRp53(1–251)F bicistronic construct as well (Supplementary Figure S8B and Supplementary Table S8C). To validate the RNA-interference results, two stable cell lines were established by puromycin selection. H1299-S3 cells stably expressed the shRNA for SMAR1, while H1299-NS cells expressed a non-targetting shRNA; IRES-mediated expression of p53 and Δ40p53 was lesser in H1299-S3 cells (Figure 2c). We also checked p53 and Δ40p53 levels on overexpressing the pcDNA p53 mammalian vector (5'UTR+cDNA; Supplementary Figure S9A) and 14A (Supplementary Figure S9B) in H1299-NS and H1299-S3 cells, in the presence and absence of DNA-damaging agent doxorubicin; results suggest that steady-state levels of both isoforms depend on SMAR1 levels. Interestingly, SMAR1 levels were also induced on DNA damage. However, it should be noted that SMAR1 can interact with full-length p53 in its TAD1 and positively regulate its stability37 and hence the levels of the full-length isoform will depend on both translation and stability.
Figure 2.
SMAR1 knockdown inhibits p53 IRES function. (a) Western blot of H1299-cell extracts co-transfected with pGFP-hp-p53-5'UTR-cDNA plasmid (14A, shown in schematic), pSV40-β-galactosidase plasmid (transfection and cap-dependent translation control) and non-targetting (Nsp) or SMAR1 siRNA. Inset shows fold change in β-galactosidase activity in each of the experimental conditions (1–6), n=3. (b) Dual-luciferase reporter assay for relative IRES activity from pRp53(1–251)F plasmid, represented by the Fluc/Rluc ratio, on SMAR1 knockdown in H1299 cells, n=9. Immunoblot in inset shows knockdown of SMAR1 in same lysates. (c) Puromycin-selected H1299 cells with stable non-targetting (NS) or SMAR1 (S3) shRNA expression were transfected with pGFP-hp-p53-5'UTR-cDNA plasmid. Western blot was done 48 h post-transfection. (d) Caspase 3/7 activation assay in H1299 cells following SMAR1 knockdown in the presence of pGFP hp-p53-5'UTR cDNA (14A plasmid). Puro, puromycin (2 μg/ml, 24 h) used as positive control for caspase induction and apoptosis. Experiment performed 48 h post-transfection, n=3. (e) Δ40p53 target 14-3-3σ mRNA levels as measured by qRT-PCR in H1299-NS and H1299-S3 cells transfected with pGFP-hp-p53-5'UTR(A135T)-cDNA (14A-M2) plasmid, n=9. (f) Western blot of H1299-NS- and H1299-S3-cell extracts that were transfected with pGFP-hp-p53-5'UTR-cDNA (14A) or pGFP-hp-p53-5'UTR(A135T)-cDNA (14A-M2) plasmid. Proteasomal degradation inhibitor MG132 (10 μM) was added 1 h before harvesting. Western blot was done on same gel and membrane; results are shown at same exposure
Caspase 3/7 activation was lessened even in the presence of p53 and Δ40p53 if SMAR1 was knocked down for 48 h (Figure 2d). The reduction in Δ40p53 expression due to SMAR1 knockdown also impaired its ability to induce transcription of downstream target genes. 14-3-3σ was recently reported as a preferential Δ40p53 target gene.5, 14 Expressing Δ40p53 from the 14A-M2 plasmid resulted in a 70% increase in 14-3-3σ mRNA in H1299-NS cells (Figure 2e). However, SMAR1 knockdown greatly compromised the induction of 14-3-3σ mRNA with only about 17% increase in the H1299-S3 cells, suggesting that SMAR1 is required for Δ40p53-mediated transactivation (Figure 2e).
We posed the question that if proteasomal degradation was inhibited by MG132 treatment (60 min) in H1299-S3 cells, will that alleviate SMAR1-knockdown-mediated repression of p53 and Δ40p53 expression. We used two constructs for this study (Figure 2f): one that would express both isoforms (14A), while another that would express only Δ40p53 (14A-M2). In either case, expression of p53 isoforms would be solely IRES mediated. On MG132 treatment after SMAR1 knockdown, there was no de-repression in the expression of p53 isoforms from either construct, suggesting that the role of SMAR1 is not at the level of protein degradation (Figure 2f). SMAR1 is known to bind to full-length p53 at 14–16 amino acids that reside within TAD-I.37 Since, SMAR1 knockdown decreased p53 as well as Δ40p53 levels expressed from different constructs and Δ40p53 lacks TAD-I, SMAR1 seems to have functions that are independent of TAD-I interaction also. A plausible explanation is that SMAR1 is indeed essential for IRES-dependent translation of p53 and Δ40p53.
p53-MDM2 protein interaction is well documented in the scientific literature. Another protein SMAR1 was found to form a ternary complex with p53-MDM2.37 As a proof of concept that SMAR1 homologs can bind p53 RNA, 6X-His-tagged mouse SMAR1 (BANP) was purified (Supplementary Figure S10A) and found to interact with p53 (1–251) IRES RNA in direct as well as competition RNA-protein UV-crosslinking assay (Supplementary Figures S10B and D), under native conditions in RNA-EMSA (Supplementary Figure S10E) and also with the 1–134 and 135–251 regions of p53 IRES RNA in filter-binding assays (Supplementary Figure S10F).
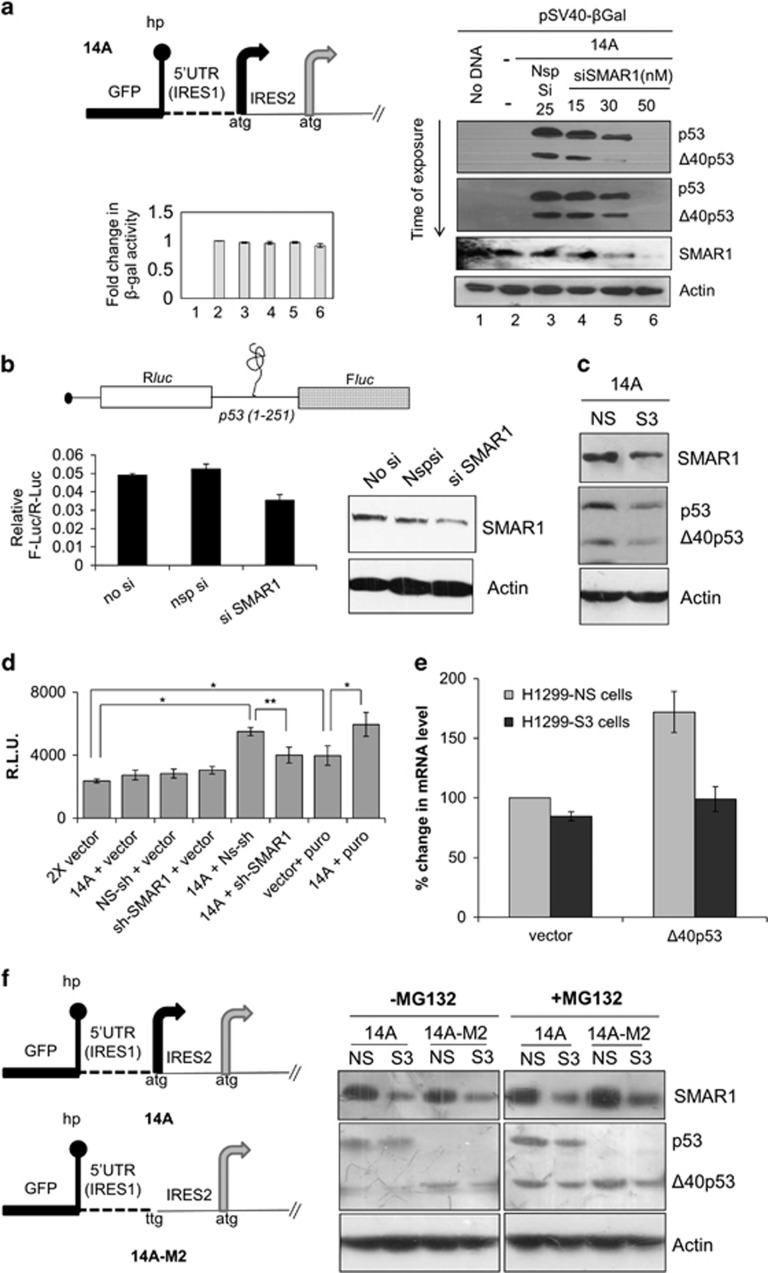
Intracellular redistribution of SMAR1 on glucose deprivation
Surprisingly, SMAR1 was seen to be redistributed in H1299 cells on glucose deprivation in immunofluorescence experiments. H1299 cells were starved of glucose for 4, 8 and 30 h, the same points in time when relative IRES activities of p53 mRNA increase. By 8 h, the nuclear SMAR1 was remarkably depleted with increase in cytoplasmic levels as compared to control cells at this time point (Figure 3a). However, by 30 h control cells and glucose-starved cells had nearly similar amounts of nuclear SMAR1 but there was more SMAR1 in the cytoplasm for the latter (Figure 3a). SMAR1 is a predominantly nuclear protein and undergoes cytoplasmic to nuclear relocalization under genotoxic stress.41 FACS studies demonstrated that glucose-deprivation-driven intracellular redistribution of SMAR1 in the cytoplasm occurred without any cues from cell-cycle changes, as the cell-cycle profiles at 4, 8 and 30 h, for control and experimental monolayer cultures were nearly identical, asynchronous and devoid of any cell-cycle arrest (Supplementary Figure S11). PTB is a very well-established ITAF for p53 IRESs and under the experimental conditions, it failed to show nuclear-cytoplasmic relocalization like SMAR1 under glucose deprivation (Figure 3b).
Figure 3.
Localization of SMAR1 and PTB after glucose deprivation (a) Intracellular redistribution of SMAR1 on glucose deprivation. Anti-SMAR1 antibody was used in indirect immunofluorescence assay on asynchronous H1299 cells starved of glucose for 4, 8 and 30 h and compared to unstarved cells at same time points. Dashed and solid arrows indicate cytoplasmic and nuclear localization of SMAR1, respectively. Fluorescently labeled secondary antibody was used and visualized by Zeiss LSM microscope at x60 objective. SMAR1 is shown in green and DAPI (nucleus) in red, whereas SMAR1 and DAPI merge is shown in yellow. (b) PTB does not relocalize to the cytoplasm on glucose deprivation. Anti-PTB antibody was used in indirect immunofluorescence assay on asynchronous H1299 cells starved of glucose for the indicated time points. Fluorescently labeled secondary antibody was used and visualized by Zeiss LSM microscope at x60 objective. PTB is shown in green and DAPI (nucleus) in blue, whereas PTB and DAPI merge is shown in cyan
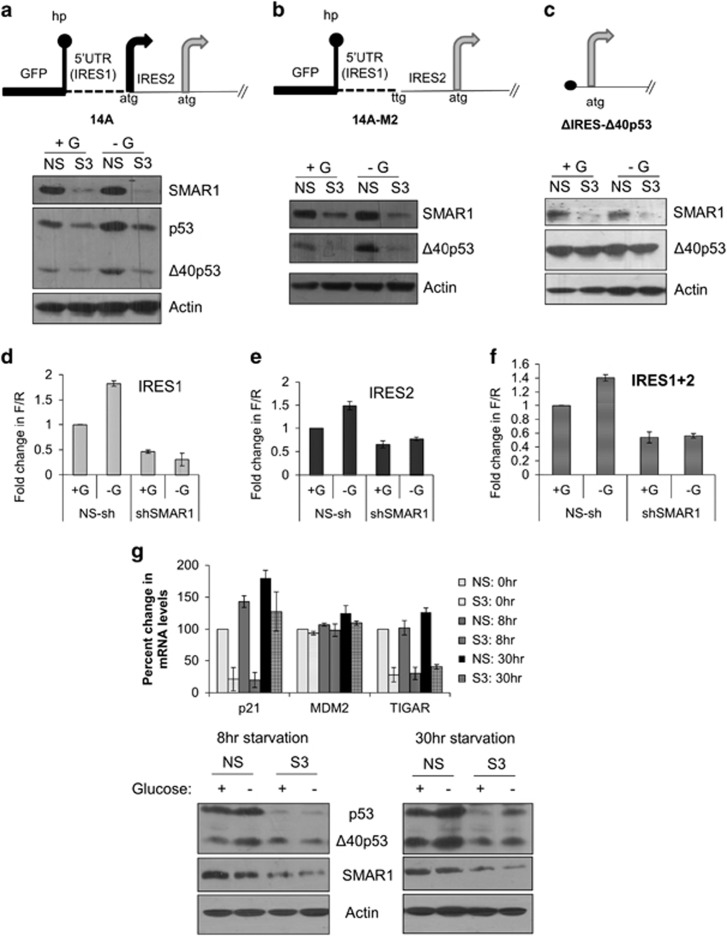
Increased expression of p53 and Δ40p53 under glucose deprivation needs SMAR1
H1299-NS and H1299-S3 cells were transfected with 14A plasmid and these cells were glucose starved for 8 h. As expected, IRES-dependent expression of p53 and Δ40p53 were lesser in H1299-S3 cells compared to H1299-NS cells (Figure 4a). Interestingly, glucose-starvation-mediated induction of the isoform levels was remarkably abrogated in the H1299-S3 cells indicating that SMAR1 is essential for such a translational induction. For studying the effect of glucose deprivation on Δ40p53 alone, a similar experiment was performed with 14A-M2 plasmid and the results corroborated the essentiality of SMAR1 for glucose-starvation-mediated induction in the synthesis of Δ40p53 (Figure 4b). Confirming our hypothesis, there was no decrease in Δ40p53 levels on SMAR1 knockdown or induction on glucose deprivation when this isoform alone was expressed in a cap-dependent manner without any IRES (ΔIRES-Δ40p53 plasmid; Figure 4c). Similarly, dual-luciferase experiments with bicistronic plasmids carrying the first (1–134) or second (135–251) IRES or with (1–251) IRES in the intercistronic region confirmed that SMAR1 is necessary for induction of IRES activity when cells are starved of glucose (Figures 4d–f; Supplementary Table S12). The expression of VEGF IRES was relatively increased on glucose deprivation, however, the expression from VEGF (Supplementary Figure S15) and HIF1α IRESs (Supplementary Figure S16) was seen to be independent of SMAR1 knockdown. Steady-state mRNA levels of three p53 transcription targets, p21, Mdm2 and TIGAR were checked in the NS and S3 cells under glucose deprivation (Figure 4g). There was an induction of all three mRNAs in NS cells but not in S3 cells, except for p21 at 30 h of starvation in the latter. Immunoblots for concomitant p53, Δ40p53 and SMAR1 levels are also shown (Figure 4g).
Figure 4.
Increased IRES-dependent expression of p53 and Δ40p53 and downstream activation of target mRNAs under glucose deprivation is SMAR1 dependent. (a–c) Western blot analysis of H1299-NS- and H1299-S3-cell extracts transfected with pGFP-hp-p53-5'UTR-cDNA plasmid (14A in schematic; a), pGFP-hp-p53-5'UTR(A135T)-cDNA plasmid (14A-M2 in schematic; b) or ΔIRES-Δ40p53 plasmid (c) that were glucose starved (−G) for 8 h before harvesting, compared with unstarved (+G) cells. (d–f) Dual-luciferase reporter assay for pRp53(1–134)F (IRES1, d), pRp53(135–251)F (IRES2, e) and pRp53(1–251)F (IRES1+2, f) bicistronic constructs transfected in H1299-NS and H1299-S3 cells that were glucose starved (−G) for 8 h before harvesting, compared with unstarved (+G) cells. Fold change in IRES activity (Fluc/Rluc) shown with this ratio unit normalized for unstarved H1299-NS cells, n=6. (g) Quantitative PCR for levels of p53-target mRNAs p21/Cip1 (CDK-interacting protein1), Mdm2 (murine double minute 2) and TIGAR (TP53-induced glycolysis and apoptosis regulator), assayed after 0, 8 and 30 h of glucose deprivation of H1299-NS and H1299-S3 cells transfected with pGFP-hp-p53-5'UTR-cDNA (14A) plasmid, n=3. Representative immunoblots depicting SMAR1, p53 and Δ40p53 levels are shown
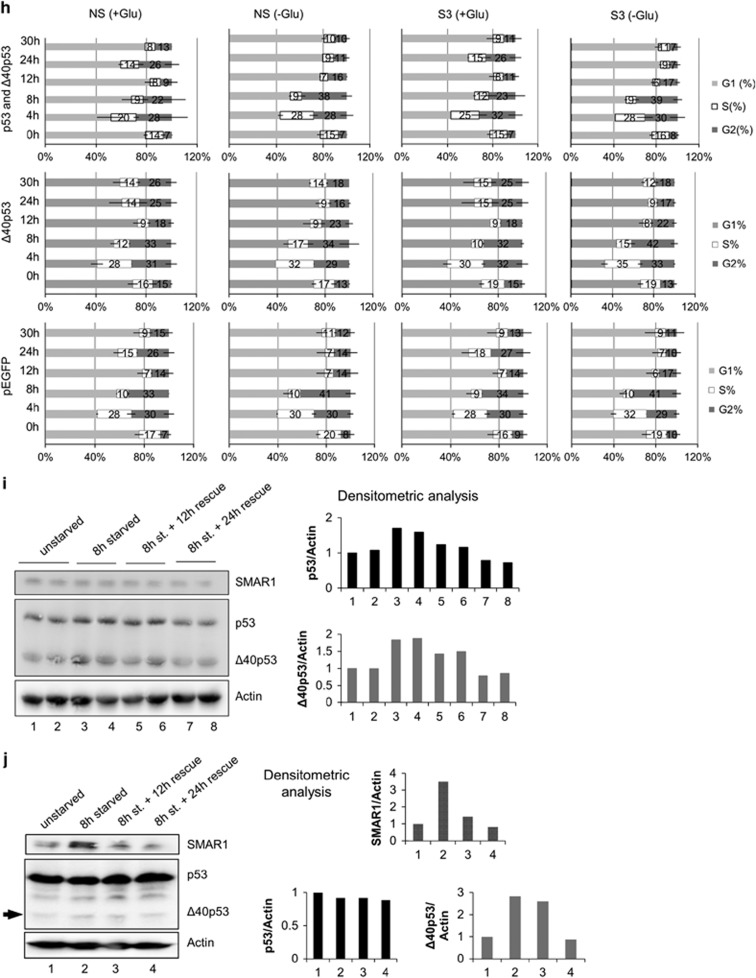
In A549 cells, changes in Δ40p53 levels reflect cap-independent-IRES-mediated translation of this isoform.3, 14 Owing to glucose deprivation, there was an increase in Δ40p53 levels at 30 h and this was confirmed with two different antibodies (Figure 5a). p53 mRNA levels do not change significantly over this time period (Supplementary Figure S17A). In A549 (Figure 5b) and HCT116 cells (Figure 5c), mRNA levels of p21 (a preferential p53 target) and SFN (a preferential Δ40p53 target) increases significantly after 30 h of glucose depletion. In A549 as well as in hepatic HepG2 cells, steady-state level of SMAR1 protein was found to be increased on glucose deprivation (Figure 5d). However, when SMAR1 was depleted using siRNA, Δ40p53 expression was almost abrogated on glucose starvation (Figure 5e). This indicates that SMAR1 is necessary for Δ40p53 expression under such conditions. The ex vivo interaction of p53 IRES was addressed by quantitative immunoprecipitation experiments. From A549 cells, RNA-protein complexes were immunoprecipitated with anti-SMAR1 antibody. The ITAF PTB, which was shown to bind p53 IRESs,3 served as a positive control (Figure 5f, anti-PTB bars). RT-qPCR analysis of the SMAR1-immunoprecipitate-associated RNA showed the presence of the RNA corresponding to 1–251 IRES, over and above than that from RNA-protein complexes immunoprecipitated by the IgG-isotype antibody, in non-starved cells (Figure 5f). There was an apparent increase in the amount of RNA pulled down by anti-SMAR1 antibody from glucose-starved A549 cells (Figure 5f). A similar experiment was performed in p53-null H1299 cells, which were transfected with GFP-hp-5'UTR-p53-bicistronic RNA (Figure 5g). There was a clear increase in IRES RNA that was pulled down with SMAR1 in conditions of glucose deprivation as compared to normal glucose (Figure 5g, anti-SMAR1 bars). Together, these results demonstrate that SMAR1 can interact with the p53 (1–251) IRES in cells and such association apparently increases on glucose deprivation. We next investigated cell-cycle progression (Figure 5h) after double-thymidine block in NS or S3 cells transfected with pEGFP (expresses only EGFP)-, 14A (construct expresses GFP, p53α and Δ40p53α)- or 14A-M2 (construct expresses GFP and Δ40p53α) and followed up with glucose starvation. Our results suggest that after 8 h of glucose deprivation in pEGFP transfected cells there is an apparent increase in 4n (and the intermediate 2n–4n) population after 8 h and the reverse at 24 h, irrespective of whether SMAR1 is knocked down or not. Interestingly this change at 8-h deprivation is absent in Δ40p53α-transfected H1299-S3 cells in glucose deprivation, preliminarily suggesting a correlation of Δ40p53α and S-phase duration in glucose deprivation that is contingent on SMAR1 levels.
Figure 5.
Physiological relevance of glucose deprivation on SMAR1-p53 IRES association. (a) Western blot of A549-cell extracts after indicated duration of glucose starvation, blotted with CM1 (left panel) or Pab240 antibody (right panel). (b,c) Quantitative PCR of p21, mdm2 and SFN mRNA levels normalized to GAPDH in A549 cells (b) and HCT116 cells (c) following 0, 8 and 30 h of glucose deprivation. (d) Western blot of A549- and HepG2-cell extracts showing the expression pattern of SMAR1 in a time-dependent manner post-glucose deprivation. Glucose was deprived for indicated time in hours and the cells were harvested at the given time points. (e) Western blot analysis of A549-cell extracts transfected with non-targetting (Nsp) or SMAR1 siRNA for 96 h, followed by glucose starvation for 30 h. Δ40p53 levels are indicated by an arrow. (f) Reverse transcriptase-quantitative PCR analysis of RNA extracted from ribonucleoprotein complexes immunoprecipitated from non-starved or 8-h glucose-starved A549 cells using anti-SMAR1, anti-PTB antibodies or IgG-isotype control. Primers corresponding to 1–251 region of p53 RNA were used. Results represented as fold change over and above RNA immunoprecipitated from non-starved cells with IgG control antibody. (g) Reverse transcriptase-quantitative PCR analysis of RNA extracted from ribonucleoprotein complexes immunoprecipitated from non-starved or 8-h glucose-starved H1299 cells transfected with GFP-hp-5'UTR-p53-bicistronic mRNA using anti-SMAR1, anti-PTB antibodies or IgG-isotype control. Primers corresponding to 1–251 region of p53 RNA were used. Results represented as fold change over and above RNA immunoprecipitated from non-starved cells with IgG control antibody. (h) FACS analysis of cell populations in G1, S or G2-M phases in H1299-NS and H1299-S3 cells. The cells were transfected with 14A (top row), 14A-M2 (middle row) or pEGFP (bottom row). Cell cycle was arrested by double-thymidine treatment, then cells were glucose starved for 0, 4, 8, 12, 24 and 30 h and harvested at these time points with corresponding unstarved control cells. (i) Glucose replenishment (rescue) for 12 and 24 h following 8 h of glucose deprivation in H1299 cells transfected with pGFP-hp-p53-5'UTR-cDNA plasmid. Western blots depict SMAR1, p53, Δ40p53 and actin levels. Graphs represent lane-wise densitometric analysis. (j) Glucose replenishment (rescue) for 12 and 24 h following 8 h of glucose deprivation in A549 cells. Western blots depict endogenous p53 and Δ40p53 (shown by an arrow) as well as SMAR1 and actin levels. Graphs represent lane-wise densitometric values. (k) Quantitative PCR for levels of p53-target mRNAs p21/Cip1 (CDK-interacting protein1), SFN (stratifin alias 14-3-3σ), Bax (Bcl-2 associated X), TIGAR (TP53-induced glycolysis and apoptosis regulator), PIDD (P53-induced DNA damage) and Mdm2 (murine double minute 2) in H1299 cells transfected with pGFP-hp-p53-5'UTR-cDNA plasmid, starved of glucose for 8 h (8 h) and then rescued by glucose replenishment for 12 h (8 h/12 h) and 24 h (8 h/24 h). Results represented as fold change in mRNA levels compared to non-starved, transfected cells, n=3
Next, we investigated whether deprivation-induced induction of p53 and Δ40p53 levels is reversible. After 8 h of glucose deprivation, H1299 (Figure 5i) and A549 cells (Figure 5i) were replenished with a medium containing glucose. In H1299 cells, IRES-driven expression of p53 isoforms from 14A plasmid returned to normal after 24 h of rescue with replenished glucose (Figure 5i). In A549 cells, endogenous levels of Δ40p53 (IRES driven) and SMAR1 increased on deprivation and returned to basal levels on 24 h of rescue (Figure 5j). The steady-state-mRNA levels of six p53 transcription targets were checked in H1299 cells transfected with pGFP-hp-p53-5'UTR cDNA (Figure 5k). There was an increase in levels of all the mRNAs, except Mdm2 on 8 h of glucose deprivation. However, on rescue, levels of p21 and SFN (cell-cycle arrest) as well as Bax and PIDD (pro-apoptotic) decreased; whereas TIGAR (glycolysis and apoptosis regulator) remained high after 8 h of rescue and decreased only after 24 h of rescue. Interestingly, Mdm2 levels were not affected significantly until 24 h of rescue, when it decreased significantly compared to non-starvation (Figure 5k).
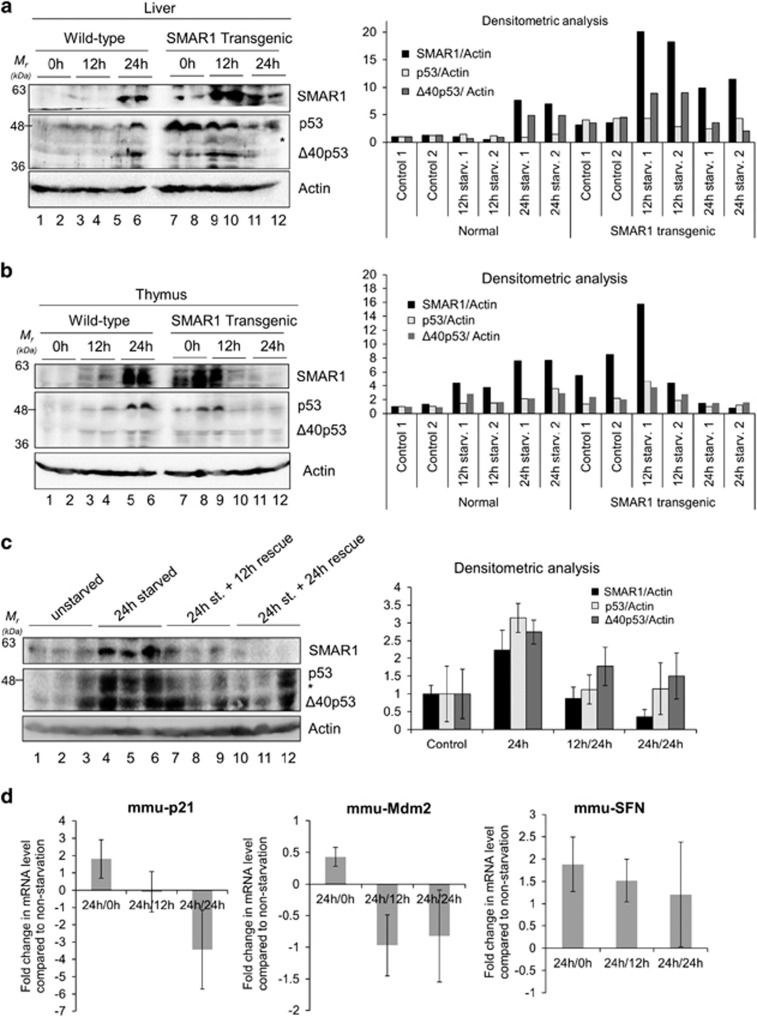
The effect of starvation on p53 and Δ40p53 was investigated in vivo. Wild-type as well as SMAR1 transgenic mice were starved for 0, 12 and 24 h. In the liver, there was a marked increase in SMAR1 levels 24 h post deprivation (Figure 6a, lanes 5 and 6) that corresponded to an increase in the levels of p44 protein (mouse Δ40p53). This was also evident in the thymus (Figure 6b, lanes 5 and 6). In the liver from SMAR1 transgenic mice, the increase in SMAR1 was more evident 12 h post deprivation with a corresponding increase in Δ40p53 at this time point (Figure 6a, lanes 9 and 10). The results, however, were more heterogeneous in the thymus from these SMAR1 transgenic mice (Figure 6b, lanes 7–12). A rescue experiment was performed in WT mice, wherein after 24 h of starvation, these were fed ad libitum for a further 12 and 24 h. Liver lysates from 12 and 24 h ‘rescued' mice showed decrease in the levels of SMAR1, p53 and, to a lesser extent, p44 (mouse Δ40p53) protein levels compared to starved mice. The mRNAs of p53 targets, p21 and Mdm2 displayed corroborating changes in their levels, returning to normal (non-starved) levels after 24 h of rescue. Blood glucose levels in both the wild-type and SMAR1 transgenic mice drop by about 20 and 40% after 12 and 24 h of starvation, respectively and are restored to normal levels after 12 h of feeding (Supplementary Figure S18).
Figure 6.
Starvation induces levels of SMAR1, p53 and Δ40p53 in vivo and this effect is reversible on withdrawal of starvation. Western blot of tissue extracts of liver (a) or thymus (b) from wild-type and SMAR1 transgenic mice at 0, 12 and 24 h of starvation, in duplicates, probed for SMAR1 (upper panel), p53 and Δ40p53 (middle panel) and actin (lower panel). Graphs show lane-wise densitometric analysis of SMAR1, p53 and Δ40p53 levels normalized to actin, n=4. ‘*' represents non-specific band. (c) Rescue experiment on starvation withdrawal for 12 and 24 h following 24 h of dietary starvation. Western blot of tissue extracts of the liver from wild-type mice probed for SMAR1 (upper panel), p53 and Δ40p53 (middle panel) and actin (lower panel). Graphs show lane-wise densitometric analysis of SMAR1, p53 and Δ40p53 levels normalized to actin, n=3. ‘*' represents non-specific band. (d) Quantitative PCR for levels of p53-target mRNAs p21/Cip1 (CDK-interacting protein1), SFN (stratifin alias 14-3-3σ) and Mdm2 (murine double minute 2) in wild-type mice liver, starved of glucose for 12 h (12 h) and then rescued by starvation withdrawal for 12 h (12 h/12 h) and 24 h (12 h/24 h). Results represented as fold change in mRNA levels compared to liver from non-starved mice, n=3. mmu: Mus musculus
Discussion
During translational control of p53 mRNA, levels of full-length p53 are predominantly regulated by cap-dependent translation initiation and IRES functions more as a back-up in stress, when cap-dependent translation declines. However, for Δ40p53 such regulation is solely IRES dependent. We report here that glucose deprivation in cultured mammalian cells induces p53 IRES activities. Earlier studies have shown that glucose starvation induces p53 protein levels in an AMP-activated protein kinase-dependent manner.42, 43 In addition, it was shown that PPAR gamma coactivator 1 alpha (PGC-1α) binds to p53 and modulates its transactivation function on glucose starvation.44 p53 IRESs are known to be inducible by ER and genotoxic stress, oncogene overexpression and G2-M arrest. The current study is the first to report that p53 IRESs are sensitive to any form of nutrient depletion. Both the IRES modules of p53 mRNA were seen to be induced on glucose starvation and higher steady-state levels of p53 and Δ40p53 could be abrogated on treatment with translation-blocker cycloheximide even in glucose depletion, further confirming the role of IRES-mediated translation in p53 mRNA under such stress. There is also a greater turnover of p53 isoforms on glucose deprivation, evident on cycloheximide treatment also. The striking feature was a complete abrogation of induced p53 isoform levels on glucose deprivation because of arrested translation with cycloheximide treatment, hence for the current study we focussed on IRES-mediated enhancement of translation of p53 isoforms on glucose deprivation. Glucose starvation has been observed as a form of ER stress because similar to this stress, depleting glucose in a medium of cultured cells results in increased eIF2α phosphorylation.39 p53-MDM2 protein interactions are well documented in the scientific literature.40, 45 MDM2 protein also binds to the coding region of p53 IRES RNA and promotes translation of the full-length p53 and the alternatively translated product Δ40p53.12, 35 p53 is also known to have RNA re-annealing properties, binding to the 5′UTR of its own mRNA,34 as well as of Cdk446 and FGF2.47, 48 Recent reports showed that SMAR1 forms a ternary complex with p53-MDM2, in which MDM2 binds to residues 17–26 of p53 and SMAR1 binds to residues 14–16 of p53, with a simultaneous interaction of SMAR1 and MDM2.37
We report that SMAR1 is able to bind to p53 (1–251) IRES directly and specifically in the in vitro studies; ex vivo immunoprecipitation experiments serve in confirming this association. SMAR1 was identified as a novel DNA-binding protein from murine thymocyte expression library screen that was associated with matrix attachment region on enhancer regions for genes encoding T-cell receptor β chain in murine CD4+CD8+ double-positive thymocytes.49 SMAR1 has been implicated in maintaining cellular homeostasis and is a global regulator/modulator of gene expression.37, 38, 50, 51, 52, 53, 54 Our recent study showed that SMAR1 is directly linked to translation of both p53 and Δ40p53 in normal as well as glucose-deprived conditions.14-3-3σ was demonstrated as a preferential transcriptional target of Δ40p53.5, 14 The downstream effects of Δ40p53 induction on target gene activation, and the resulting biological outcome, have been previously explored.55, 56, 57 In the current study, using Δ40p53-mediated transactivation as a model, we showed that SMAR1 is important for IRES-dependent Δ40p53 induction, as well as for the consequent elevation of 14-3-3σ mRNA (Figure 2e). Therefore, activation of p53 IRES by SMAR1 is likely to contribute to Δ40p53 function, probably more than full-length p53, as translational induction of the shorter isoform has to be through IRES and hence ITAF mediated. Also, p53-mediated transactivation was found to be lesser in case of SMAR1-knockdown cells, largely in both non-starved and starved conditions (Figure 2e). Nuclear-cytoplasmic relocalization of ITAFs is essential for cellular IRES function.58, 59, 60 We report in the current study a glucose-dependent relocalization of SMAR1, but not PTB, in cultured cells, highlighting the novelty of SMAR1-mediated control in stress. In addition, rescue experiments both ex vivo and in vivo show that the induction of p53 isoform levels on nutrient deprivation is reversible, with these returning to basal levels on nutrient replenishment. The changes in p53 transactivation target mRNAs also largely corroborate this finding.
Deregulations in translational control contribute to each step of cellular transformation and tumor progression, nutrient starvation being a very common stress for the cells located inside non-vascularized tumors.61 Activation of p53 IRESs on glucose deprivation provides a new regulatory aspect to the field of translational control of p53 mRNA, suggesting critical function of the p53 isoforms in these conditions in terms of cell survival and stress tolerance.
Materials and Methods
Plasmid constructs
Dual-luciferase constructs of the p53 (1–251) IRES, first (1–134) IRES, second (135–251) IRESs,3, 8 VEGF and HIF1α IRESs (kind gifts from Professor Greg Goodall, Centre of Cancer Biology, Australia and Professor Annapoorni Rangarajan, MRDG, IISc) and HCV IRES62 were used in quantitative bicistronic assays. WWP luciferase (a kind gift from Professor Kumaravel Somasundaram, IISc) was used for measuring p53-dependent transactivation, CMV-Renilla-luciferase (Promega, Madison, WI, USA) constructs were used to normalize transfection efficiency. GFP-p53-bicistronic constructs used were pGFP-hp-p53-5'UTR-cDNA-containing initiator ATGs for both p53 and Δ40p53 (a kind gift from Professor Robin Fahraeus, INSERM, France), pGFP-hp-p53-5'UTR-(A252G/T253C/G254T) cDNA with a functional initiator ATG for p53 only, and pGFP-hp-p53-5'UTR(A135T) cDNA with a functional initiator ATG for Δ40p53 only. pSV40-β-galactosidase (Promega, Madison, WI, USA) was used as a control for transfection efficiency as well as cap-dependent translation. shRNA-mediated knockdown of SMAR1 was through pGIPZ-shSMAR1(S3) construct, pGIPZ-NS construct expressed non-targeting shRNA. ΔIRES-Δ40p53 construct was used for cap-dependent expression of Δ40p53 (a kind gift from Professor Robin Fahraeus, INSERM, France and Professor Adi Kimchi, Weizmann Institute of Science, Israel). pCMV-3XFLAG-SMAR1 was used as described earlier.53
Cell lines, transfections, glucose deprivation and drug treatment
H1299, HCT116-p53+/+, A549 and HepG2 cells were maintained in DMEM (Sigma-Aldrich, St. Louis, MO, USA) with 10% fetal bovine serum (Biological Industries, Beit Haemek, Israel). For luciferase assays or western blots, 70% confluent monolayer of H1299 or A549 were transfected with various bicistronic luciferase constructs or cDNA plasmids using Lipofectamine 2000 (Invitrogen, Life Technologies, Carlsbad, CA, USA) in Opti-MEM (Invitrogen). Four hours later, the medium was replaced with DMEM (with antibiotic) and 10% FBS. At the desired time point, the cells were harvested and processed as required. For glucose deprivation, cells were washed twice in DMEM-minus glucose and maintained in the same medium for the desired duration. For establishing cell populations with stable expression of SMAR1 or non-targeting shRNA, corresponding pGIPZ DNA constructs were transfected into H1299 cells. After 24 h, 2-μg/ml puromycin was added in the medium and this concentration was maintained for the next 48 h. Then the pooled-puromycin-resistant cells were passaged two times in 1-μg/ml puromycin and SMAR1 knockdown validated by immunoblotting. For all subsequent experiments, pooled-antibiotic-resistant H1299-NS and H1299-S3 cells were maintained in 1-μg/ml puromycin. For DNA-damage induction, H1299-NS and S3 cells were treated with 2-μM doxorubicin for 16 h.
siRNA transfection
The employed siRNA sequence against SMAR1 was 5′-UAACCCUGAGAUGCGGGUA-3′. As a control for silencing, a siCONTROL non-targeting siRNA #5 (Dharmacon, Thermo Scientific, Waltham, MA, USA) was used in the experiments in a similar manner. Co-transfection of siRNA with various plasmid constructs was performed in monolayer H1299, HCT116-p53+/+ or A549 cells using Lipofectamine 2000 (Invitrogen) transfection reagent in Opti-MEM (Invitrogen). Forty-eight or 96 h post-transfection the cells were harvested and the extracts were used for dual-luciferase assays or for western blot analysis.
Western blot analysis
Protein concentrations of the extracts were assayed by Bradford (Bio-Rad, Hercules, CA, USA) and equal amounts of cell extracts were resolved in SDS-PAGE, 12% and transferred to nitrocelluose membrane (Sigma-Aldrich). Samples were then analyzed by western blotting using rabbit-raised CM-1 antibody (kind gift from Professor Robin Fahraeus, INSERM), anti-SMAR1 antibody (Abcam, Cambridge, UK), mouse-raised DO1 and Pab240 antibodies (Santa Cruz Biotechnology, Dallas, TX, USA) or rabbit-raised anti-GFP antibody (IMG5127, Imgenex, Bhubaneswar, Odisha, India) followed by secondary antibody (horseradish peroxidase-conjugated anti-rabbit IgG; Sigma-Aldrich). Mouse-monoclonal anti-β-actin antibody (Sigma-Aldrich) was used as a control for equal loading of total cell extracts. Antibody complexes were detected using the Immobilon Western systems (EMD Millipore, Billerica, MA, USA).
Dual-luciferase assay
Renilla- and firefly-luciferase activities were measured using the dual-luciferase reporter assay system (Promega), according to the manufacturer's protocol, by using TD20/20 Luminometer (Turner BioSystems, Promega).
Beta-galactosidase assay
RIPA lysates from pSV40-β-galactosidase-transfected cells were used for the colorimetric assay. To 0.1-M sodium phosphate buffer (82% 0.1-M Na2HPO4 and 18% 0.1-M NaH2PO4), total-protein normalized volumes of RIPA lysates were added along with 0.1-M MgCl2, 4.5-M β-mercaptoethanol and 4-mg/ml o-nitrophenyl-β,d-galactopyranoside substrate; reactions were incubated in 37 °C for 30 min. After quenching with 1-M sodium carbonate solution A420 nm was measured in a spectrophotometer.
Caspase assay
Eight thousand H1299 cells were seeded per well in 96-well plates 16 h prior to transfection. These were transfected with a total of 15-ng DNA per well and the medium was changed after 6 h. After 48 h, 50 μl of caspase 3/7 Glo reagent (Promega) was added per well, incubated at room temperature for 3 h and then luminescence was measured in Turner BioSystems plate reader using the corresponding protocol.
RNA isolation and real-time PCR
Total RNA was isolated from H1299, A549 or HCT116 cells using TRI reagent (Sigma-Aldrich) as per manufacturer's protocol, treated with 10 units of DNase per sample and extracted by phenol-chloroform method. For RNA isolation from mice livers, a part of the organ was suspended in TRI reagent (Sigma-Aldrich) and 200-μg/ml glycogen, homogenized by handheld Down's homogenizer (Sigma-Aldrich) by 50 strokes on ice and then manufacturer's protocol was followed as usual. For real-time PCR, first-strand cDNA was synthesized using oligo-(dT)18 primer (Thermo Scientific) using RevertAid RT enzyme (Thermo Scientific) from 2-μg RNA. Reaction was set up with 2 μl of 1 : 5 diluted cDNA using DyNAmo qPCR kit (Thermo Scientific) that uses SYBR Green chemistry as per manufacturer's instructions on ABI Prism 7900HT machine (Applied Biosystems, Life Technologies) or ABI ViiA7 machine (Applied Biosystems, Life Technologies). Data were analyzed using SDSv2.3 software (Applied Biosystems, Life Technologies) or ViiA7 RUO software (Applied Biosystems, Life Technologies). The results were calculated by the comparative ΔΔ Ct method. The primer sequences are as follows:
p53 Forward: 5′-TGGGCTTCTTGCATTCTGG-3′, p53 reverse: 5′-GCTGTGACTGCTTGTAGATGGC-3′ p21 forward: 5′-CCTCAAATCGTCCAGCGACCTT-3′ p21 reverse: 5′-CATTGTGGGAGGAGCTGTGAAA-3′ BAX forward: 5′-GCCCTTTTGCTT CAGGGTTT-3′ BAX reverse: 5′-TCCAATGTCCAGCCCATGAT-3′ PIDD forward: 5′-CGAGCCCTCTGACACGGT-3′ PIDD reverse: 5′-AGAAGGACACCTGGCCCC-3′ SFN forward (14-3-3σ): 5′-TGAGAACTGGACAGTGGCAG-3′ SFN (14-3-3σ) Reverse: 5′-GAGGAAACATGGTCACACCC-3′ MDM2 forward: 5′-ATCTTGGCCAGTATATT ATG-3′ MDM2 reverse: 5′-GTTCCTGTAGATCATGGTAT-3′ TIGAR forward: 5′-CTGACTGAAACTCGCTAAGG-3′ TIGAR reverse: 5′-CAGAACTAGCAGAGGA GAGA-3′ actin forward: 5′-TCACCCACACTGTGCCCATCTACGA-3′ actin reverse: 5′-TGAGGTAGTCAGTCAGGTCCC-3′ mmu-p21 forward: 5′-CGGTGGAACTTTGACTTCGT-3′ mmu-p21 reverse: 5′-GAGTGCAAGACAGCGACAAG-3′ mmu-Mdm2 forward: 5′-AGATTCCAGCTTCGGAACAA-3′ mmu-Mdm2 reverse: 5′-ACACAATGTGCTGCTGCTTC-3′ mmu-SFN forward: 5′-GAAACCTGCTTTCCGTAGCTTA-3′ mmu-SFN reverse: 5′-TCTGAGCTCGGTCTCTACCTTC-3′ mmu-Actin forward: 5′-CGGTTCCGATGCCCTGAGGCTCTT-3′ mmu-actin reverse: 5′-CGTCACACTTCATGATGGAATTGA-3′.
Immunofluorescence staining
For immunofluorescence staining, H1299 cells were grown on coverslips for 14 h followed by glucose deprivation for 4, 8, 17 and 30 h. At each time point, cells on coverslips were washed twice with 1x PBS and fixed by 4% formaldehyde at room temperature for 20 min. After permeabilization by 0.1% Triton X-100 for 2 min at room temperature, cells were incubated with 4% BSA at 37 °C for 1 h followed by incubation with rabbit-raised anti-SMAR1 (Abcam) antibody or mouse-raised anti-PTB antibody (Calbiochem, White House Station, NJ, USA) for 16 h at 4 °C and then detected by Alexa-488-conjugated anti-rabbit or Alexa-633-conjugated anti-mouse secondary antibody for 30 min (Invitrogen). Images were acquired using Zeiss confocal microscope and image analysis was done using LSM Image browser (Zeiss, Oberkochen, Germany).
FACS analysis
For FACS analysis, the asynchronous monolayer of H1299 cells was trypsinized, washed with 1x PBS and fixed in 70% ethanol at 4 °C. Fixed cells were treated with 50 μg of RNase A (Thermo Scientific) at 37 °C for 60 min and then stained with 20-ng/ml propidium iodide (Sigma-Aldrich) at 37 °C for 30 min. Stained cells were analyzed in FACSCantoII (BD Biosciences) cell analyzer using FACS Diva 2.0 software (BD Biosciences).
Immunoprecipitation of RNP complexes in vivo
GFP-hp-5'UTR-p53-bicistronic mRNA was synthesized from XhoI linearized pGFP-hp-p53-5'UTR-cDNA plasmid using RiboMax kit (Promega) as per manufacturer's instructions. H1299 cells at 85–90% confluence 16 h post-seeding were transfected with these RNAs. After 4 h, no-glucose DMEM with 10% FBS was added to the experimental set of cells. After another 8 h, cells were lysed with polysome lysis buffer (100-mM KCl, 5-mM MgCl2, 10-mM HEPES at pH 7.0, 0.05% NP-40, 1-mM DTT, 100-U/ml RNasin and 1% protease inhibitor cocktail). Similarly, untransfected A549 cells were glucose deprived for 8 h, no deprivation was employed for control A549 cells and after 8 h cells were lysed with polysomal lysis buffer. Supernatants were pre-cleared with Fast-Flow protein-G sepharose beads (Sigma-Aldrich) for 30 min at room temperature. Pre-cleared lysates containing equal total protein (500 μg) were incubated with anti-SMAR1, anti-PTB- (Calbiochem) or rabbit IgG (Sigma-Aldrich) saturated protein-G sepharose beads overnight at 4 °C. RNP complexes conjugated to antibody-bound beads were spun down at 8000 r.p.m. for 2 min, washed four times in ice-cold polysomal lysis buffer and spun down similarly. RNP complexes were precipitated followed by proteinase-K (Promega) treatment for 30 min at 55 °C. RNA was extracted using TRI reagent (Sigma-Aldrich) as per manufacturer's protocol and treated with DNase-I (Promega) at 37 °C for 20 min. Finally, with phenol-chloroform-precipitated RNA, RT-PCR was performed using IRES-specific reverse primer and RevertAid reverse transcriptase (Thermo Scientific). cDNA was used for quantitative PCR using IRES-specific primers and PCR products resolved in 2% agarose gel. The primer sequences are as follows: p53-10-27-F-IP: 5′-ACCGTCCAGGGAGCAGGT-3′, p53-238-251-R-IP: 5′-TGCTTGGGACGGCA -3′. Input was normalized using the values obtained in RNA isolated from 10% input lysate.
In vivo experiments in mice
A total of 24 eight-week-old female C57BL/6 mice (12 WT and 12 transgenic; for SMAR1)63 were employed in the experiment. Three time points were used, for example, 0 h (control), 12 h (of starvation) and 24 h (of starvation). At each time point four mice per experimental set were killed and the required tissues (liver and thymus) were isolated. For the rescue experiment, a total of 12 WT mice were employed, three each for control, 24-h starvation, 24-h starvation/12-h rescue and 24-h starvation/24-h rescue batches. The tissues were first washed in ice-cold 1x PBS and were finely minced using a scissor and forceps, on ice. In total, 30 mg of the minced tissue sample was suspended in 1x RIPA buffer and was subjected to sonication (15 s/15 s; pulse/pause) at high intensity for a total time of 15 min in a sonication machine (Bioruptor, Diagenode, Denvile, NJ, USA). The samples were then centrifuged at 12 000 r.p.m. for 30 min. The supernatant was collected and processed for immunoblotting. For blood glucose level determination, wild-type (C57BL/6) and SMAR1 transgenic (SMAR1Tg) mice were used to set up the experiment. Blood was collected by heart puncture and the serum was isolated. The serum glucose levels were measured by GOD-POD method using a kit (Spinreact, Girona, Spain). The absorbance was measured at 505 nm in an ELISA plate reader. The levels were then calculated with reference to the standard sample. The graph represents the data of four independent experiments.
In vitro transcription
For mSMAR1-binding analysis of p53 (1–251) IRES, the respective bicistronic plasmid DNA was PCR amplified using T7 promoter sequence-tagged p53 forward primer and specific reverse primer. Amplified DNA was purified using PCR-purification kit (Qiagen, Venlo, The Netherlands) as per manufacturer's instructions. As per manufacturer's instructions the α-32P-UTP-labeled RNA probe of 1–251 IRES RNA was synthesised with T7 RNA polymerase and 10 μCi/μl of α-32P-UTP (NEN, Perkin Elmer, Waltham, MA, USA) and used for UV-crosslinking assays. In addition, 40 units of RNase inhibitor (Promega) were included in each reaction in order to inhibit the activity of contaminating nucleases.
Protein purification
The expression of recombinant mouse SMAR1 was induced by 1-M IPTG for 16 h in Escherichia coli (BL21 DE3) cells transformed with pET28b-SMAR1. His-tagged protein was purified using Ni2+-nitrilotriacetic acid-agarose (Qiagen) under non-denaturing conditions and eluted with 500-mM imidazole.
UV-induced crosslinking of proteins and RNA
UV-induced crosslinking was carried out as described elsewhere.3 Briefly, α-32P-UTP-labeled 1–251 IRES RNA probe was allowed to form complexes with recombinant mSMAR1 at 30 °C for 25 min in 1x RNA-binding buffer and then irradiated with a handheld UV lamp for 20 min. The mixture was treated with 30-μg RNase A (Sigma-Aldrich) at 37 °C for 30 min. The protein-nucleotidyl complexes were electrophoresed on SDS-PAGE, 10% and analyzed by phosphorimaging.
Electrophoretic mobility shift assay
Conditions for EMSA have been previously described.64 α 32P-UTP-labeled p53 1–251 IRES (25 fmol) was incubated with 300 and 600 ng of His-SMAR1 protein at 30 °C for 30 min. Loading dye was added and the protein-nucleotidyl complex was resolved on a 4% (60 : 1) polyacrylamide gel at 4 °C.
Filter-binding assay
The α-32P-labeled corresponding p53 RNAs (1–251 or 1–134 or 135–251 regions of p53 IRES) were incubated with increasing concentrations of purified mSMAR1 at 30 °C for 20 min in RNA-binding buffer (5-mM HEPES at pH 7.6, 2- mM KCl, 2-mM MgCl2, 3.8% glycerol, 2-mM DTT and 0.1-mM EDTA) and loaded onto nitrocellulose filters equilibrated again with RNA-binding buffer. The filters were washed four times, dried and the counts retained were measured in a scintillation counter. The graph was plotted with protein concentration (micromolar concentrations) on x-axis and percentage of RNA bound as the percentage of counts retained on y-axis.
Statistical analysis
The data were expressed as mean±S.D. Statistical significance was determined using two-sided Student's t-test. The criterion for statistical significance was P<0.05 (*) or P<0.005 (**).
Acknowledgments
We thank Professor Robin Fahraeus (INSERM, France) and Profesor Adi Kimchi (Weizmann Institute of Science, Israel) for sharing various p53 cDNA constructs, Professor Greg Goodall (Centre of Cancer Biology, Australia) and Professor Annapoorni Rangarajan (MRDG, IISc) for sharing VEGF and HIF1α bicistronic constructs and Professor K Somasundaram (MCB, IISc) for sharing the WWP-luciferase construct. We also thank the FACS facility (Division of Biological Sciences, IISc) for the acquisition of the flow-cytometric data. We also thank the present and past SD lab-members for critical discussion of the work. DK, AS and AD were supported by pre-doctoral fellowships from Council of Scientific and Industrial Research, India. This work was supported by a grant from the Department of Biotechnology, India to Sa.D. Sa.D and SC thank JC Bose fellowship from DST, Government of India.
Author Contributions
DK contributed to conception and design of studies, data acquisition, analysis and interpretation and writing the article; AK was involved in generation of mutant constructs and interpretation of corresponding data, co-acquisition of quantitative PCR data, co-acquistion and analysis of FACS data; AD was responsible for acquisition, analysis and interpretation of in vivo data; RL contributed to acquisition, analysis and interpretation of in vitro binding data; AS was involved in acquisition, analysis and interpretation of immunofluorescence data; PR contributed to acquisition, analysis and interpretation of data from cell lines expressing endogenous p53 isoforms; Sr.D was responsible for acquisition of in vitro binding data; and SC and Sa.D contributed to conception and design of studies, data analysis and interpretation and writing the article.
Glossary
- IRES
internal ribosome entry site
- ER
endoplasmic reticulum
- ITAFs
IRES trans-acting factors
- R-Luc
Renilla luciferase
- F-Luc
firefly luciferase
- CHX
cycloheximide
- TAD I
transactivation domain
- 14A
pGFP-hp-p53-5'UTR-cDNA
- 14A-M1 plasmid
pGFP-hp-p53-5'UTR-(A252G/T253C/G254T)-cDNA
- 14A-M2 plasmid
pGFP-hp-p53-5'UTR(A135T)-cDNA
- mSMAR1
mouse SMAR1
- SMAR1Tg
SMAR1 transgenic
- EMSA
electrophoretic mobility shift assay
- DOXO
doxorubicin
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Oren
Supplementary Material
References
- 1Candeias MM. The can and can't dos of p53 RNA. Biochimie 2011; 93: 1962–1965. [DOI] [PubMed] [Google Scholar]
- 2Halaby MJ, Yang DQ. p53 translational control: a new facet of p53 regulation and its implication for tumorigenesis and cancer therapeutics. Gene 2007; 395: 1–7. [DOI] [PubMed] [Google Scholar]
- 3Grover R, Ray PS, Das S. Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle 2008; 7: 2189–2198. [DOI] [PubMed] [Google Scholar]
- 4Belin S, Beghin A, Solano-Gonzalez E, Bezin L, Brunet-Manquat S, Textoris J et al. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS One 2009; 4: e7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Bourougaa K, Naski N, Boularan C, Mlynarczyk C, Candeias MM, Marullo S et al. Endoplasmic reticulum stress induces G2 cell-cycle arrest via mRNA translation of the p53 isoform p53/47. Mol Cell 2010; 38: 78–88. [DOI] [PubMed] [Google Scholar]
- 6Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, Montanaro L et al. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res 2010; 70: 6026–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Grover R, Sharathchandra A, Ponnuswamy A, Khan D, Das S. Effect of mutations on the p53 IRES RNA structure: implications for de-regulation of the synthesis of p53 isoforms. RNA Biol 2011; 8: 132–142. [DOI] [PubMed] [Google Scholar]
- 8Ray PS, Grover R, Das S. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep 2006; 7: 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Marcel V, Ghayad SE, Belin S, Therizols G, Morel AP, Solano-Gonzalez E et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 2013; 24: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Su H, Xu T, Ganapathy S, Shadfan M, Long M, Huang TH et al. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene 2014; 33: 1348–1358. [DOI] [PubMed] [Google Scholar]
- 11Khan D, Sharathchandra A, Ponnuswamy A, Grover R, Das S. Effect of a natural mutation in the 5' untranslated region on the translational control of p53 mRNA. Oncogene 2013; 32: 4148–4159. [DOI] [PubMed] [Google Scholar]
- 12Candeias MM, Malbert-Colas L, Powell DJ, Daskalogianni C, Maslon MM, Naski N et al. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol 2008; 10: 1098–1105. [DOI] [PubMed] [Google Scholar]
- 13Sharathchandra A, Lal R, Khan D, Das S. Annexin A2 and PSF proteins interact with p53 IRES and regulate translation of p53 mRNA. RNA Biol 2012; 9: 1429–1439. [DOI] [PubMed] [Google Scholar]
- 14Weingarten-Gabbay S, Khan D, Liberman N, Yoffe Y, Bialik S, Das S et al. The translation initiation factor DAP5 promotes IRES-driven translation of p53 mRNA. Oncogene 2014; 33: 611–618. [DOI] [PubMed] [Google Scholar]
- 15Kim DY, Kim W, Lee KH, Kim SH, Lee HR, Kim HJ et al. hnRNP Q regulates translation of p53 in normal and stress conditions. Cell Death Differ 2013; 20: 226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 2005; 123: 49–63. [DOI] [PubMed] [Google Scholar]
- 17Chen J, Guo K, Kastan MB. Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA. J Biol Chem 2012; 287: 16467–16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Wedeken L, Singh P, Klempnauer KH. Tumor suppressor protein Pdcd4 inhibits translation of p53 mRNA. J Biol Chem 2011; 286: 42855–42862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Zhang M, Zhang J, Chen X, Cho SJ, Chen X. Glycogen synthase kinase 3 promotes p53 mRNA translation via phosphorylation of RNPC1. Genes Dev 2013; 27: 2246–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Marr MT 2nd, D'Alessio JA, Puig O, Tjian R. IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev 2007; 21: 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Bruhat A, Cherasse Y, Chaveroux C, Maurin AC, Jousse C, Fafournoux P. Amino acids as regulators of gene expression in mammals: molecular mechanisms. Biofactors 2009; 35: 249–257. [DOI] [PubMed] [Google Scholar]
- 22Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 2003; 11: 619–633. [DOI] [PubMed] [Google Scholar]
- 23Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol 2002; 22: 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Fernandez J, Yaman I, Mishra R, Merrick WC, Snider MD, Lamers WH et al. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J Biol Chem 2001; 276: 12285–12291. [DOI] [PubMed] [Google Scholar]
- 25Gaccioli F, Huang CC, Wang C, Bevilacqua E, Franchi-Gazzola R, Gazzola GC et al. Amino acid starvation induces the SNAT2 neutral amino acid transporter by a mechanism that involves eukaryotic initiation factor 2alpha phosphorylation and cap-independent translation. J Biol Chem 2006; 281: 17929–17940. [DOI] [PubMed] [Google Scholar]
- 26Oltean S, Banerjee R. A B12-responsive internal ribosome entry site (IRES) element in human methionine synthase. J Biol Chem 2005; 280: 32662–32668. [DOI] [PubMed] [Google Scholar]
- 27Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell 2000; 11: 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Cullen PJ, Sprague GF Jr.. Glucose depletion causes haploid invasive growth in yeast. Proc Natl Acad Sci USA 2000; 97: 13619–13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Gilbert WV, Zhou K, Butler TK, Doudna JA. Cap-independent translation is required for starvation-induced differentiation in yeast. Science 2007; 317: 1224–1227. [DOI] [PubMed] [Google Scholar]
- 30Sherrill KW, Lloyd RE. Translation of cIAP2 mRNA is mediated exclusively by a stress-modulated ribosome shunt. Mol Cell Biol 2008; 28: 2011–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk RG. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol 1999; 1: 190–192. [DOI] [PubMed] [Google Scholar]
- 32Wang G, Miskimins R, Miskimins WK. Regulation of p27(Kip1) by intracellular iron levels. Biometals 2004; 17: 15–24. [DOI] [PubMed] [Google Scholar]
- 33Cho S, Kim JH, Back SH, Jang SK. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol Cell Biol 2005; 25: 1283–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J 1995; 14: 4442–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Yin Y, Stephen CW, Luciani MG, Fahraeus R. p53 stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol 2002; 4: 462–467. [DOI] [PubMed] [Google Scholar]
- 36Grover R, Candeias MM, Fahraeus R, Das S. p53 and little brother p53/47: linking IRES activities with protein functions. Oncogene 2009; 28: 2766–2772. [DOI] [PubMed] [Google Scholar]
- 37Pavithra L, Mukherjee S, Sreenath K, Kar S, Sakaguchi K, Roy S et al. SMAR1 forms a ternary complex with p53-MDM2 and negatively regulates p53-mediated transcription. J Mol Biol 2009; 388: 691–702. [DOI] [PubMed] [Google Scholar]
- 38Sinha S, Malonia SK, Mittal SP, Singh K, Kadreppa S, Kamat R et al. Coordinated regulation of p53 apoptotic targets BAX and PUMA by SMAR1 through an identical MAR element. EMBO J 2010; 29: 830–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 2001; 7: 1165–1176. [DOI] [PubMed] [Google Scholar]
- 40Picksley SM, Vojtesek B, Sparks A, Lane DP. Immunochemical analysis of the interaction of p53 with MDM2—fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene 1994; 9: 2523–2529. [PubMed] [Google Scholar]
- 41Singh S, Sreenath K, Pavithra L, Roy S, Chattopadhyay S. SMAR1 regulates free radical stress through modulation of AKR1a4 enzyme activity. Int J Biochem Cell Biol 2010; 42: 1105–1114. [DOI] [PubMed] [Google Scholar]
- 42Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 2005; 18: 283–293. [DOI] [PubMed] [Google Scholar]
- 43Okoshi R, Ozaki T, Yamamoto H, Ando K, Koida N, Ono S et al. Activation of AMP-activated protein kinase induces p53-dependent apoptotic cell death in response to energetic stress. J Biol Chem 2008; 287: 3979–3987. [DOI] [PubMed] [Google Scholar]
- 44Sen N, Satija YK, Das S. PGC-1alpha, a key modulator of p53, promotes cell survival upon metabolic stress. Mol Cell 2011; 44: 621–634. [DOI] [PubMed] [Google Scholar]
- 45Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992; 69: 1237–1245. [DOI] [PubMed] [Google Scholar]
- 46Miller SJ, Suthiphongchai T, Zambetti GP, Ewen ME. p53 binds selectively to the 5' untranslated region of cdk4, an RNA element necessary and sufficient for transforming growth factor beta- and p53-mediated translational inhibition of cdk4. Mol Cell Biol 2000; 20: 8420–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Gonzalez-Herrera IG, Prado-Lourenco L, Pileur F, Conte C, Morin A, Cabon F et al. Testosterone regulates FGF-2 expression during testis maturation by an IRES-dependent translational mechanism. FASEB J 2006; 20: 476–478. [DOI] [PubMed] [Google Scholar]
- 48Gonzalez-Herrera IG, Prado-Lourenco L, Teshima-Kondo S, Kondo K, Cabon F, Arnal JF et al. IRES-dependent regulation of FGF-2 mRNA translation in pathophysiological conditions in the mouse. Biochem Soc Trans 2006; 34: 17–21. [DOI] [PubMed] [Google Scholar]
- 49Chattopadhyay S, Kaul R, Charest A, Housman D, Chen J. SMAR1, a novel, alternatively spliced gene product, binds the scaffold/matrix-associated region at the T cell receptor beta locus. Genomics 2000; 68: 93–96. [DOI] [PubMed] [Google Scholar]
- 50Jalota A, Singh K, Pavithra L, Kaul-Ghanekar R, Jameel S, Chattopadhyay S. Tumor suppressor SMAR1 activates and stabilizes p53 through its arginine-serine-rich motif. J Biol Chem 2005; 280: 16019–16029. [DOI] [PubMed] [Google Scholar]
- 51Singh K, Sinha S, Malonia SK, Bist P, Tergaonkar V, Chattopadhyay S. Tumor suppressor SMAR1 represses IkappaBalpha expression and inhibits p65 transactivation through matrix attachment regions. J Biol Chem 2009; 284: 1267–1278. [DOI] [PubMed] [Google Scholar]
- 52Pavithra L, Singh S, Sreenath K, Chattopadhyay S. Tumor suppressor SMAR1 downregulates cytokeratin 8 expression by displacing p53 from its cognate site. Int J Biochem Cell Biol 2009; 41: 862–871. [DOI] [PubMed] [Google Scholar]
- 53Rampalli S, Pavithra L, Bhatt A, Kundu TK, Chattopadhyay S. Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol Cell Biol 2005; 25: 8415–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54Sinha S, Malonia SK, Mittal SP, Mathai J, Pal JK, Chattopadhyay S. Chromatin remodelling protein SMAR1 inhibits p53 dependent transactivation by regulating acetyl transferase p300. Int J Biochem Cell Biol 2012; 44: 46–52. [DOI] [PubMed] [Google Scholar]
- 55Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev 2004; 18: 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56Ohki R, Kawase T, Ohta T, Ichikawa H, Taya Y. Dissecting functional roles of p53 N-terminal transactivation domains by microarray expression analysis. Cancer Sci 2007; 98: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57Powell DJ, Hrstka R, Candeias M, Bourougaa K, Vojtesek B, Fahraeus R. Stress-dependent changes in the properties of p53 complexes by the alternative translation product p53/47. Cell Cycle 2008; 7: 950–959. [DOI] [PubMed] [Google Scholar]
- 58Spriggs KA, Bushell M, Mitchell SA, Willis AE. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ 2005; 12: 585–591. [DOI] [PubMed] [Google Scholar]
- 59Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene 2008; 27: 1033–1035. [DOI] [PubMed] [Google Scholar]
- 60Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 2011; 10: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol 2013; 5: pii: a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62Ray PS, Das S. Inhibition of hepatitis C virus IRES-mediated translation by small RNAs analogous to stem-loop structures of the 5'-untranslated region. Nucleic Acids Res 2004; 32: 1678–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63Kaul-Ghanekar R, Majumdar S, Jalota A, Gulati N, Dubey N, Saha B et al. Abnormal V(D)J recombination of T cell receptor beta locus in SMAR1 transgenic mice. J Biol Chem 2005; 280: 9450–9459. [DOI] [PubMed] [Google Scholar]
- 64Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc 2007; 2: 1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.