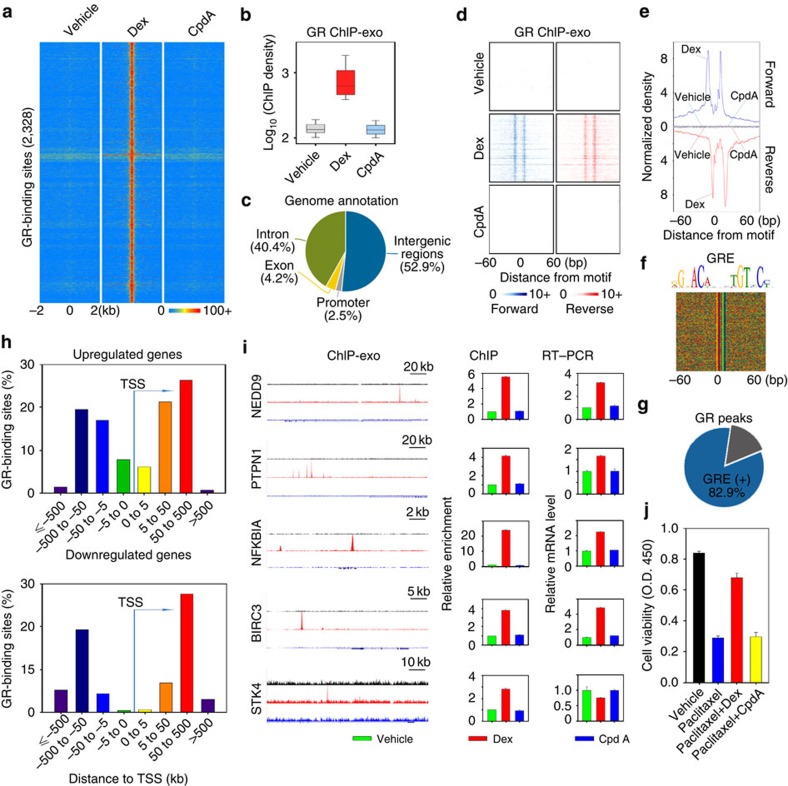
Figure 4. Precise definition and characterization of the GRE recognized by Dex-liganded GR in MDA-MB-231 cells.
(a) Heat maps show the signal intensity of GR binding in MDA-MB-231 cells treated with vehicle, 100 nM Dex or 10 μM CpdA for 1 h. The number (2,328) indicates Dex-responsive GR locations. (b) A box plot shows ChIP-exo signal densities in Dex-responsive GR locations in cells treated with vehicle, Dex or CpdA. (c) Classification of specific Dex-responsive GR-binding locations based on annotation. (d) Raw tags distribution and (e) aggregated tag density over Dex GRE under different treatment conditions is shown on the forward (blue) and reverse (red) strands, separately. (f) Dex GRE is shown and the sequences are presented in the same order as in d. (g) Percentage of Dex-responsive GR-binding locations containing Dex GRE. (h) The distribution of Dex GR binding around the transcription start site (TSS) of upregulated (upper panel) and downregulated (lower panel) GR target genes are shown. (i) Left panel: UCSC genome browser views of ChIP-exo sequencing data at five gene loci. Y-scale is the same for each gene locus. Colours represent different treatment conditions: vehicle (green), red (Dex) and blue (CpdA). Middle panel: ChIP validation of GR ChIP-exo data. Cells were treated with vehicle, 100 nM Dex or 10 μM CpdA for 1 h and GR ChIP was performed. For the PTPN1 locus, ChIP was performed to validate the binding of GR to the highest binding peak. Right panel: messenger RNA levels of five genes were also examined by reverse transcriptase–PCR (RT–PCR) assays. ChIP and RT–PCR data are the mean of triplicates±s.d. (j) Cells were treated with vehicle, 100 nM paclitaxel, 100 nM paclitaxel/100 nM Dex or 100 nM paclitaxel/10 μM CpdA and cell proliferation was determined on day 4 using the WST-1 assay. The data are the mean of triplicates±s.d.