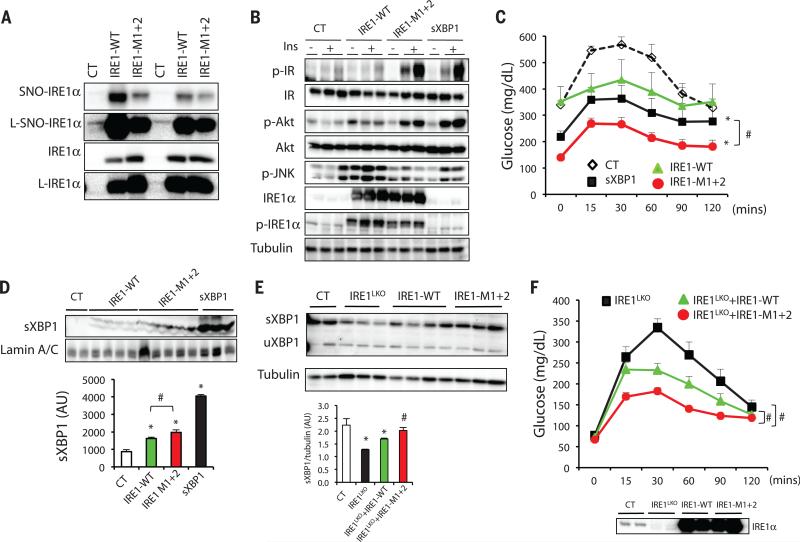
Fig. 4. Effects of IRE1αa S-nitrosylation on insulin action and glucose homeostasis.
(A to D) ob/ob mice were transduced with adenovirus containing sXBP1, WT IRE1α or nitrosylation-resistant IRE1-M1+2. (A) S-Nitrosylation of IRE1α in primary hepatocytes from these mice. The nitrosylation state of IRE1α was examined by biotin switch assay. L, longer exposure time. Data are representative of two individual mouse cohorts. (B) Hepatic insulin action and UPR status. N = 6 to 7, data are representative of two individual cohorts of mice. (C) Glucose tolerance test. All data are presented as means ± SEM, with statistical analysis of the area under the curve (AUC) performed by two-way ANOVA with post hoc Bonferroni test. Asterisk (*) indicates statistical significance compared with control, and # indicates differences between sXBP1 and IRE1-M1+2 expressing mice. Data are representative of two individual cohorts of mice [n = 8 (10-week-old mice)]. (D) sXBP1 expression in the nuclear fraction of liver, with densitometric analysis shown at the bottom of panel. Asterisk (*) indicates statistical significance compared with control, and # indicates statistically significant differences between IRE1-WTand IRE1-M1+2 (one-way ANOVA followed by post hoc Tukey's test). Data are representative of two individual mouse cohorts. (E and F) IRE1α-floxed mice were transduced with Ad-LacZ (Control) or Ad-Cre to delete IRE1α (IRE1LKO), followed by expression of IRE1-WT or the IRE1-M1+2 variant. (E) Hepatic expression of sXBP1 and uXBP1 in the liverof these mice. Quantification of sXBP1/tubulin ratio is shown below. Asterisk (*) indicates statistical significance compared with control, and # indicates statistically significant differences between IRE1-WT and IRE1-M1+2 (one-way ANOVA followed by post hoc Tukey's test). Data are representative of two independent sets of mice. (F) Glucose tolerance test of the mice shown in (E). (Bottom) The IRE1α expression levels are shown [n = 8 (9-week old mice)]. Data are presented as means ± SEM, with statistical analysis of AUC performed by two-way ANOVA with post hoc Bonferroni test. Number sign (#) indicates statistically significant differences comparing IRE1LKO+IRE-M1+2 with IRE1LKO+IRE1-WT, as well as IRE1LKO+IRE1-WT with IRE1LKO. Data are representative of two individual cohorts of mice. *P < 0.05; #P < 0.05.