Abstract
The lack of live human brain cells for research has slowed progress toward understanding the mechanisms underlying autism spectrum disorders (ASDs). A human model using reprogrammed patient somatic cells offers an attractive alternative as it captures a patient’s genome in relevant cell types. Despite the current limitations, the disease-in-a-dish approach allows for progressive time-course analyses of target cells, offering a unique opportunity to investigate the cellular and molecular alterations before symptomatic onset. Understanding the current drawbacks of this model is essential for the correct data interpretation and extrapolation of conclusions applicable to the human brain. Innovative strategies for collecting biological material and clinical information from large patient cohorts are important for increasing the statistical power that will allow for the extraction of information from the noise resulting from the variability introduced by reprogramming and differentiation methods. Working with large patient cohorts is also important for understanding how brain cells derived from diverse human genetic backgrounds respond to specific drugs, creating the possibility of personalized medicine for ASD.
Keywords: human-specific disease modeling, human induced pluripotent stem cells, human neurons, brain, drug screening, autism spectrum disorders
Introduction
Autism Spectrum Disorder (ASD) is a lifelong developmental disability that is mainly characterized by difficulties in social communication and the presence of focused repetitive or stereotyped behaviors and appears within the first three years of life(1). Because different etiologies can generate a similar behavioral outcome, many disorders with autistic features are grouped under the ASD umbrella.
Family history and twin studies suggest that these disorders share genetic roots in at least some cases (2, 3). The mounting evidence suggests that heritable and de novo genetic variations play a significant role, but these studies have also reported striking genetic heterogeneity(4-6). Disorders such as Fragile X, Rett syndrome (RTT) and Timothy syndrome are caused by specific genetic alterations that also present neurodevelopmental and speech delays, resulting in an autistic phenotype. Although these syndromic forms are no longer clinically grouped under ASD, these disorders have provided useful insights into sporadic or idiopathic (non-syndromic) forms of autism. Current genomic efforts to discover novel causative variants have focused on small chromosomal deletions or duplications in the form of copy number variations (CNVs) measured by the genotyping of large numbers of individuals(7, 8). CNVs were found in cadherins and protocadherins, implicating the neuronal cell adhesion pathway in ASD or the ubiquitin-proteasome system, which regulates synaptic attributes such as neurotransmitter release and synaptic vesicle recycling(9). Other studies have found that genes with rare CNV defects interfere with neurodevelopmental pathways by affecting the maturation and function of glutamatergic synapses that may be disrupted in ASD(7, 10). Recent studies integrating ASD candidate genes with spatiotemporal coexpression networks have demonstrated that ASD genes converge on the transcriptional regulation in pyramidal (glutamatergic) cortical neurons during mid-fetal human development(11, 12).
Neuropathological imaging has also provided important insights into ASD. Macrocephaly and altered brain development trajectories with early overgrowth and later normalization have been reported in some ASD patients(13). This increase in brain size during the first three years of life was shown to precede the first clinical manifestation(14-19). Several pieces of evidence suggest that accelerated brain growth in this ASD population begins prenatally and continues during the first few years of life(14, 17, 20-22). Some MRI findings correlate directly with a post-mortem analysis, such as a weight (size) increase in ASD brains at early ages, describing the neurons as more packed and a reduced number of Purkinje cells in the cerebellum(23). Subsequent studies have focused on identifying cellular abnormalities, such as increases in the number of neurons(24) and the glial density(25) in the prefrontal cortex.
The prevalence rate of ASD has dramatically risen over the years. The exact reasons for this increase remain unclear; however, the improvement in and availability of diagnosis and a legitimate increase in the rate of affected newborns may be contributing factors(26, 27). According to the CDC’s 2014 Autism and Developmental Disabilities Monitoring (ADDM) Network, approximately 1 in 68 children has been identified with an ASD in the United States. ASD is almost five times more common among males than females.
There is no cure for ASD. ASD treatment requires a strong collaboration among multiple professionals, and the cornerstone of treatment involves individualized educational interventions, including early and intensive behavioral strategies and therapies for better clinical outcomes(28, 29). The cost for this type of personalized treatment can be quite high(30). As these children mature into autistic adults, the majority do not live independently(31). Thus, the need for early diagnosis and better treatment of ASD is not only an increasing concern among scientists and physicians but also an increasing concern from an economic perspective(32). However, the human nature of ASD, with its intrinsic heterogeneity and large spectrum of clinical symptoms among patients, is a major challenge for studying ASD.
Current ASD models
Studies using several experimental models have improved our understanding of ASD. The best models consider the sophistication of the human brain within the constraints of cost and practicality. The inaccessibility to live neurons, either from post-mortem brains or living individuals, has hindered the investigation of the mechanisms underlying ASDs. Other issues associated with post-mortem analyses are similar to those associated with live imaging, such as the sample size, gender, age and heterogeneity of the disorder itself. All of the above-mentioned issues are added to the possible lack of information on the medical or drug-use history of the individuals whose brains are being studied and the differences in the methodology or statistical analyses used among the research groups. Nonetheless, useful information on the ASD pathology has been extracted from gene expression studies using postmortem brain tissues(33, 34). Furthermore, we have also gained a significant amount of knowledge of the genetics of ASD by performing genomic analyses on blood samples from affected and non-affected individuals(8, 35, 36). However, these are not ideal cell types for neuroscience experimentation because blood cells do not exhibit several of the specialized structures (for example, the synaptic machinery) found in neurons. Fetal primary human progenitor/stem cells represent an acceptable experimental ASD model, but the intrinsic difficulties in their manipulation, expansion and accessibility restrict their use(37, 38).
Finally, the inherent differences between the mouse and human genetic backgrounds(39), immune systems(40) and brain circuits(41) contribute to the challenges of using rodent models of ASD(42). With a relatively shorter evolutionary distance and a more heterogeneous genetic background than inbred laboratory mice, non-human primate models have also been used to study ASD(43). Recent efforts have also focused on the targeted genetic manipulation of non-human primates to carry alterations found in syndromic forms of ASD, but mechanistic insights from these models have not yet been reported(44, 45) and may be inaccurate because of the uniqueness of the brain transcriptional networks in modern humans(34)(46). Thus, a new human model with unlimited access to relevant cellular material could nicely complement the efforts from previous approaches.
A human pluripotent experimental model for ASD
Despite some ethical controversy, human embryonic stem cell (hESC) research has become a promising area in developmental research. For the first time, researchers can potentially explore the early stages of human development in vitro using this powerful tool to gain insights into human neurodevelopment(47). However, the progress toward understanding neurodevelopmental diseases has been hampered by the scarce availability of disease-specific hESCs carrying ASD genetic alterations in the genome. The generation of induced pluripotent stem cells (iPSCs) using human cells has been accomplished(48, 49). This breakthrough involves a relatively simple approach that uses a set of transcription factors to jump-start and reprogram the entire genetic network landscape to a pluripotent stage. In addition to overcoming ethical issues, this new technique has gained attention for its potential to generate disease-specific pluripotent stem cells with unprecedented simplicity. Human iPSCs are tempting models for understanding complex disorders with heritable and sporadic conditions, which allow researchers to recapitulate an individual’s development in the laboratory(50).
Given the uniqueness of human cognition and behavior, an in vitro human neurodevelopmental model capable of recapitulating the early stages of development could reveal specific biochemical and cellular features of our species that are difficult to reproduce in other models(37, 51). The iPSC technology also presents the potential opportunity to manipulate phenotypic alterations with candidate drugs, paving the way for future drug-screening platforms.(52).
Major roadblocks to establishing an iPSC ASD human model
The introduction of iPSCs has been a breakthrough for studying human diseases. However, understanding the limitations of this approach is important to extract the most relevant data from this model. As with all in vitro models, cells in culture are not in the same environment as the living organism. Additionally, the current culture conditions are not yet fully optimized for deriving enriched populations of disease-relevant neuronal subtypes(53-60) or glial cells(61) from human pluripotent stem cells despite recent efforts.
A recent study comparing primary human fetal progenitor cells, which were isolated directly from the developing brain, with differentiating iPSCs found that although both of these in vitro models recapitulated certain aspects of human corticogenesis and the transcriptional network related to ASD, the iPSC-derived neurons exhibited relatively lower transcriptional overlap with in vivo human development(38). The high variability reported for these experiments clearly demonstrates that different experimental protocols may affect the degree of cortical maturity that iPSC-derived neurons can achieve in culture.
Another relevant issue concerns the use of proper cellular controls. Based on previous experiences with mouse models, isogenic cell lines may represent the ultimate control. Genome-editing allows for rigorous study designs that could substantially alleviate concerns regarding potential off-target effects or other confounding effects caused by clonal sequence variation(62-65). Additionally, genome-editing technologies can be used to add reporters, biosensors and tagged proteins to easily assay biological endpoints in a more homogenous cellular population. Random X chromosome silencing during the reprogramming of female cell lines is another strategy for generating isogenic control cell lines when the target gene is located on the X chromosome(5, 66). However, the eventual loss of X chromosome inactivation that could artificially “rescue” cellular phenotypes is a concern(67). In complex diseases, in which the genetic contribution comes from multiple genetic variants, it may be advantageous to coordinate collaborative initiatives to share control lines(68).
Finally, confirming that the in vitro cellular phenotypes are relevant to the disease is currently another major obstacle for investigators in this field. Because several cellular and molecular phenotypes in a dish may not be recapitulated in postmortem brain tissues or animal models for the reasons previously discussed, confirmation in another model would definitely increase the confidence in disease relevance.
Undoubtedly, the iPSC technology still requires a series of optimization steps, which are necessary for not only the study of ASDs but also the study of any neurological disorder. Despite all these caveats and limitations, the study of ASD-derived brain cell types is already generating novel insights that should be considered with an open-mind for novel perspectives(69).
Modeling syndromic ASDs using iPSCs
We have previously used iPSCs to study Rett syndrome (RTT), a syndromic type of ASD(5) caused by alterations in the methyl-CpG-binding protein-2 (MeCP2) gene. RTT-derived cortical glutamatergic neurons exhibited several morphometric alterations and reduced excitatory synapses, leading to significant functional deficits in the RTT neuronal networks. Interestingly, the morphometric observations mirrored the data from RTT postmortem brain tissues to some extent(70). Moreover, treating the RTT-derived neurons with insulin growth factor 1 (IGF1) increased the number of synapses, rescuing the neuronal defects. IGF1 is now in clinical trials for treating RTT and potentially other ASDs(71). This outcome suggests that the iPSC-derived neurons may be helpful in the screening of future candidate drugs. Our methods have been subsequently validated and improved upon by other research groups, suggesting that this platform can be quite robust despite the potential caveats previously discussed(66, 72, 73). Since this first report, several other scientific articles showing that iPSCs can be used to model other syndromic types of autism have been published.
Fragile X syndrome (FXS) is caused by a CGG repeat expansion in the promoter region of the fragile X mental retardation 1 (FMR1) gene, which leads to hypermethylation and gene silencing(74). Initial developmental studies on FXS using human embryonic stem cells derived from blastocysts revealed that the FMR1 promoter was unmethylated and thus expressed at the pluripotent stage(75). In contrast, initial studies using FXS iPSCs showed that FMR1 was inactive because it retained the epigenetic silent status(76). A follow-up study showed that the FMR1 silent status actually varied among several clonal iPSC lines(77). Moreover, the lines with reduced FRM1 expression generated aberrant neuronal differentiation(77). Another recently reported study showed that the FXS-derived cortical neurons were aberrant neurons with reduced neuritogenesis(78). Thus, despite the initial hurdles, these reports demonstrate the potential of iPSCs to generate FXS models in a dish.
Timothy Syndrome (TS) is another rare ASD that is caused by mutations in the CACNA1C gene(79). A human iPSC model for TS has helped identify several abnormalities in the derived neurons, including defects in calcium signaling, reductions in cortical and callosal projection markers, an increased number of neurons expressing tyrosine hydroxylase and an activity-dependent dendritic retraction (80, 81). Interestingly, the increase in tyrosine hydroxylase was rescued after treatment with roscovitine, an L-type channel blocker, emphasizing the potential for a drug-screening platform using TS-derived neurons. Neuroligins, a class of genes previously implicated in ASD, were also targeted for disease modeling using reprogramming techniques(53, 82).
Finally, syndromic non-monogenetic ASDs were also modeled using human reprogramming techniques. Phelan-McDermid syndrome (PMDS), also known as 22q13.3 deletion syndrome, is caused by a loss of genes located in the 22q13 chromosomal region, which typically includes deleting SHANK3, a gene previously implicated in autism(83). The neurons generated from two PMDS patients demonstrated an altered excitatory electrophysiology and fewer synapses compared with controls. These neuronal defects were rescued by the ectopic expression of SHANK3, further supporting the causative role of SHANK3 in this syndrome(84). Maternally inherited duplications of the 15q11-q13.1 chromosomal region are also associated with ASD(85). This region includes the ubiquitin protein ligase E3A (UBE3A), which is disrupted in another neurodevelopmental disorder called Angelman syndrome(86). The transcriptional analysis of iPSC-derived neurons from five patients with CNVs in 15q11-q13.1 revealed that the chromatin structure can influence the gene expression across this region, suggesting that common molecular pathways may be disrupted in both Angelman and the duplication syndrome(87). A similar gene expression strategy was used to model CNVs in chromosome 7q11.23, a region that contains approximately 25 genes. Deletions in this region cause Williams Syndrome, whereas duplications are generally associated with ASDs(88). The authors showed that transcriptional dose-dependent dysregulation can even be detected at the iPSC stage, but it is enhanced in the differentiated cell types(89). The CNV models described here strongly suggest that the general approach is sound and reliable, thereby allowing for the discovery of important molecular pathways related to ASD pathogenesis.
Modeling non-syndromic ASDs using iPSCs
Based on the examples of syndromic ASD, it can be tentatively concluded that the iPSC-derived neurons of idiopathic or non-syndromic ASD patients can serve as an important tool for studying rare ASD variants(88, 90-93)(94, 95). However, demonstrating the causal role for rare variants will be particularly challenging because it may require an unattainable large sample size for proper statistical power(96-98).
Based on this caveat, our group has focused on the reprogramming of human dental pulp stem cells from ASD children using the deciduous tooth as a source of somatic cells. The “Tooth-Fairy Project”(99) has dramatically increased over the past years because of the use of social networks to connect with ASD families. We have been collecting dental pulp from ASD patients from Brazil and the USA, and we have a cellular and genetic ASD bank that represents part of the heterogeneity of the ASD population. Currently, we have over 3,500 ASD families listed and approximately 300 samples collected. The bank will cross-reference all the genotypes and cellular phenotypes with detailed clinical information. The bank will be useful for modeling the differences in the ASDs patients with distinct clinical outcomes using reprogramming strategies. Moreover, we believe that iPSCs together with genomic analyses, such as whole-genome sequencing, will help categorize ASDs and clarify some of the molecular pathways leading to the autistic behavior.
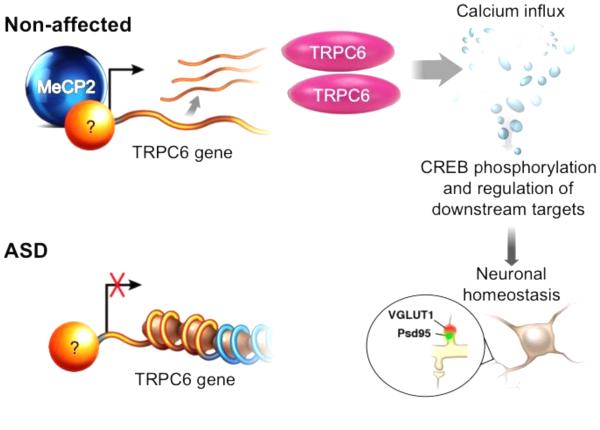
Recently, we have also made progress in modeling non-syndromic autism. Starting with the dental pulp cells isolated from one of the ASD subjects enrolled on the “Tooth-Fairy Project,” we characterized the breakpoints of a de novo balanced translocation t(3;11)(p21;q22) that disrupts the TRPC6 gene. TRPC6, a gene not previously implicated in ASD, encodes the canonical transient receptor potential 6 channel, a voltage-independent Ca2+-permeable cation channel involved in dendritic spine and excitatory synapse formation(100, 101). The biological impact of the genetic alteration in the index case and its functional relationship to the ASD etiology was evaluated in several analyses using cortical neurons derived from iPSCs. We demonstrated that a TRPC6 reduction or haploinsufficiency leads to altered neuronal development, morphology, and function(54). The observed neuronal phenotypes could then be rescued by TRPC6 complementation and by treatment with IGF1 or hyperforin, a TRPC6-specific agonist, suggesting that ASD individuals with alterations in this pathway may benefit from these drugs. We also demonstrated that the MeCP2 level affects TRPC6 expression, revealing common pathways among ASDs (Figure 1). The genetic sequencing of TRPC6 in 1041 ASD individuals and 2872 controls revealed that there are significantly more nonsynonymous mutations in the ASD population and identified loss-of-function mutations with incomplete penetrance in two patients(54). Taken together, these findings suggest that TRPC6 is a novel predisposing gene for ASD that may act in a multiple-hit model. This was the first study to use iPSC-derived human neurons to model non-syndromic ASD and illustrate the potential of modeling genetically complex sporadic diseases using these types of cells.
Figure 1.
The use of iPSCs to model ASD has already revealed new biological insights. A recent ASD predisposition gene (TRPC6) was shown to contribute to the formation of glutamatergic synapses in human neurons. Interestingly, TRPC6 gene expression can be controlled by MeCP2, a gene implicated in Rett syndrome, revealing the shared molecular pathways between syndromic and non-syndromic autisms. In non-affected individuals, MeCP2 is part of a currently uncharacterized activator complex in cortical neurons, stimulating the expression of TRPC6 that promotes calcium influx. The downstream activation pathway involves CREB-targets that regulate neuronal homeostasis, including the establishment of excitatory neurons. In a subset of idiopathic ASD individuals, TRPC6 is downregulated due to a reduced level of MeCP2 or mutations in the TRPC6 allele.
A drug-screening platform using iPSC-derived brain cell types
The lack of pharmacological treatments for the core symptoms of ASD may be a consequence of the lack of a human model. ASD pre-clinical assays are mostly based on transformed cell lines and animal models that cannot fully recapitulate the relevant cellular function because of obvious interspecies differences and differences in their pharmacokinetic properties(102, 103). Drug-screening platforms require suitable cellular phenotypes for the high-throughput instrumentation screening and a large amount of the target cell types. The current differentiation strategies can be further developed for use in scale-up methods in the near future. Morphometric measurements can be easily taken using high-content imaging software. Gene expression differences and biochemical assays are also valuable alternatives. However, functional electrophysiology studies may not be practical due to the long maturation time required in vitro. Direct cell fate conversion may be an alternative(104, 105), but the ability to scale up production and the skipping of important neurodevelopmental milestones may be a challenge(106).
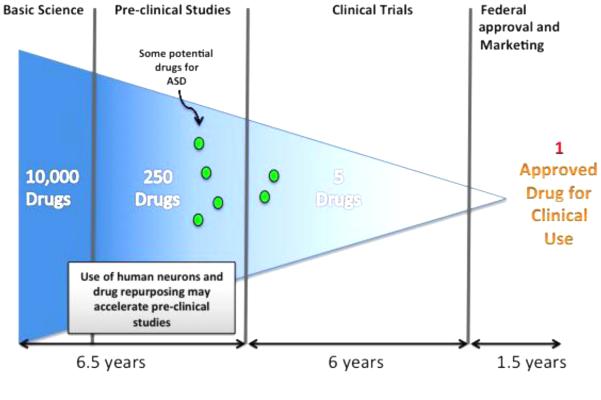
Although most of the research has focused on neurons, the transition to a high-throughput drug-screening platform has intrinsic challenges mainly because iPSC-neurons are dynamic cell types that require constant attention over long periods of time. It is notoriously difficult to produce a homogenous population of neuronal subtypes, and human neurons are not well adapted to the high-throughput format. Therefore, the use of other well-characterized populations of functional cell types that are easily adapted to high-throughput formats may provide a significant advantage for drug-screening initiatives. Strong evidence suggests that ASDs are diseases of the synapse, the interconnection through which electrical signals are transmitted from one neuron to the next in a circuit. Most of the research has focused on new therapeutics that target neurons and enhance the number of functional synapses in these diseases. However, a recent study has shown that glial cells may represent an exciting novel therapeutic target, and they should be investigated in the future. New protocols for the generation of glia will allow researchers to identify the role of these cell types in ASD. Previous studies have established that astrocytes secrete signaling molecules that stimulate the formation, pruning and function of synapses throughout the brain(107, 108). Astrocyte dysfunctions are known to be associated with many mouse models of neurological diseases, but their specific contribution to the human disease pathology has yet to be determined. It is likely that some glial cells, such as astrocytes and microglia, also contribute to the disease. By mixing different cell types in co-culture experiments, it will be possible to isolate the non-cell autonomous contribution to specific neuronal parameters, such as morphology or synaptogenesis. Finally, ideal drug libraries for ASD should consist of molecules with good brain penetration. Repurposing drugs designed for other disorders for ASD is another fascinating idea that could accelerate promising pre-clinical leads to market (Figure 2).
Figure 2.
Incorporation of iPSCs can accelerate drug discovery for ASDs. Adding iPSC-derived neurons can potentially impact the time frame for screening and marketing new drugs for ASDs. As the models become more sophisticated and the cost of iPSCs becomes reduced, this type of approach may also be used to categorize the ASD-responsive populations for more efficient treatment.
The next step: mini-brains on a chip
Drug development typically begins with a screening that tests the effects of promising molecules on 2D cell cultures and then in animal models before reaching clinical trials. However, many successful pre-clinical candidates still fail in humans. Pluripotent stem cells grown in 3D cultures can self-organize into spheres that resemble a 9-week-old developing human fetus brain, forming cerebral organoids that more accurately represent the actual conditions than the 2D cultures(109, 110). Obviously, these structures are not small-scaled brains because several components are not present or appear in different places. Nonetheless, this strategy offers a panoramic picture of brain development and how the process can go wrong. An additional step would be to help organize these cells by taking advantage of tissue engineering.
Modern tissue engineering applies concepts and techniques from engineering and biology to the problems that occur when cultivating human tissues and organs in the laboratory. Typically, this process involves creating a scaffold of natural or synthetic material, seeding this scaffold with human stem cells, which are provided with specific nutrients and a specific physical environment, that will differentiate into particular cell types and populate the structure(111). Despite the success with simple tissues, such as cartilage or skin, the reconstruction of complex organs in a dish is a major challenge. Nonetheless, the generation of human neuronal microcircuits is already a reality(112). For example, scientists have designed microfluidic chambers that allow iPSC-derived hippocampal neurons to connect with cortical neurons. Multi-electrode arrays (MEA) can be incorporated to record electrical impulses or stimulate the circuitry and investigate the connectivity of functional human networks(113). As another example, human motor neurons could connect to muscle to study neuromuscular junctions. By having the different cells functioning together, these circuits will replicate the dynamic interactions that take place in the body. Perhaps this model will be the first type of in vitro-in vivo human correlation in the neuroscience field. The MEA recordings from these neuronal assemblies can be directly correlated with the EEG recordings from patients, for example.
Obviously, the “brain on a chip” does not precisely replicate the human brain. Generally, these are plastic devices with tiny chambers embedded with living cells that are connected by microfluidic channels that control the flow of nutrients and oxygen in the system. However, breaking the brain into microcircuits has both financial and convenience advantages. A microscale version of the brain is easy to manipulate and the outcomes are more easily visualized, supporting multiple variables at the same time. The system has the potential to reduce or eliminate the dependency on animal models and to accelerate drug development, helping identify the ineffective or unsafe drugs earlier in the development process. Personalized medicine strategies will also gain from this technology. However, as the culture systems become more sophisticated, new strategies will emerge along with new challenges. Nonetheless, the effort and work required to generate these strategies is worth the time. In the future, doctors may no longer have to guess which of the several drugs and dosages would work best for a specific patient. They could simply test them all in a micro-model of the patient’s brain and select the one with the greatest efficacy and the fewest side effects.
Perspective
Human-derived cells are a powerful tool that nicely complements other models for studying ASDs. Although the iPSC technology is still in its early stage, it has demonstrated the potential ability to recapitulate the relevant neuronal defects of these diseases. The disease-in-a-dish approach could eventually incorporate data from other areas, such as systems biology, computational simulations, human brain imaging, and population genetics, to generate novel working hypotheses to be tested with more suitable cell types. Moreover, future advances in human-relevant models and technologies will generate a research approach that avoids the limitations of the species barrier. These types of initiatives can be combined with open-source interfaces that are user-friendly, allowing scientists to share key information to develop Adverse Outcome Pathways (AOPs) for autism spectrum disorders, an increasingly useful tool in toxicology. AOPs can be established for different autisms to accumulate evidence that supports the biological information underlying specific clinical symptoms. This strategy could be very useful for supporting the regulatory decision-making political proposals for autistic individuals. Finally, as the technology evolves, it will become applicable to personalized medicine, making predictions on the efficiency of certain drugs and doses in specific individuals.
Acknowledgments
I would like to thank members of my laboratory for critical comments and suggestions on this manuscript. The work in the Muotri lab is supported by grants from the California Institute for Regenerative Medicine (CIRM) TR2-01814 and TR4-06747, the National Institutes of Health through the NIH Director's New Innovator Award Program, 1-DP2-OD006495-01, the International Rett Syndrome Foundation (IRSF grant # 2915), the Humane Society International and a NARSAD Independent Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The author reports no biomedical financial interests or potential conflicts of interest.
References
- 1.Wing L, Gould J. Severe impairments of social interaction and associated abnormalities in children: epidemiology and classification. Journal of autism and developmental disorders. 1979;9:11–29. doi: 10.1007/BF01531288. [DOI] [PubMed] [Google Scholar]
- 2.Piven J, Palmer P, Jacobi D, Childress D, Arndt S. Broader autism phenotype: evidence from a family history study of multiple-incidence autism families. The American journal of psychiatry. 1997;154:185–190. doi: 10.1176/ajp.154.2.185. [DOI] [PubMed] [Google Scholar]
- 3.Ronald A, Happe F, Bolton P, Butcher LM, Price TS, Wheelwright S, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Child Adolesc Psychiatry. 2006;45:691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- 4.Garber K. Neuroscience. Autism's cause may reside in abnormalities at the synapse. Science. 2007;317:190–191. doi: 10.1126/science.317.5835.190. [DOI] [PubMed] [Google Scholar]
- 5.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook EH, Jr., Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 8.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, et al. Patches of disorganization in the neocortex of children with autism. N Engl J Med. 2014;370:1209–1219. doi: 10.1056/NEJMoa1307491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Courchesne E, Campbell K, Solso S. Brain growth across the life span in autism: age-specific changes in anatomical pathology. Brain Res. 2011;1380:138–145. doi: 10.1016/j.brainres.2010.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courchesne E, Redcay E, Morgan JT, Kennedy DP. Autism at the beginning: microstructural and growth abnormalities underlying the cognitive and behavioral phenotype of autism. Dev Psychopathol. 2005;17:577–597. doi: 10.1017/S0954579405050285. [DOI] [PubMed] [Google Scholar]
- 16.Hazlett HC, Poe MD, Gerig G, Styner M, Chappell C, Smith RG, et al. Early brain overgrowth in autism associated with an increase in cortical surface area before age 2 years. Arch Gen Psychiatry. 2011;68:467–476. doi: 10.1001/archgenpsychiatry.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Courchesne E, Karns CM, Davis HR, Ziccardi R, Carper RA, Tigue ZD, et al. Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study. Neurology. 2001;57:245–254. doi: 10.1212/wnl.57.2.245. [DOI] [PubMed] [Google Scholar]
- 18.Courchesne E, Carper R, Akshoomoff N. Evidence of brain overgrowth in the first year of life in autism. JAMA. 2003;290:337–344. doi: 10.1001/jama.290.3.337. [DOI] [PubMed] [Google Scholar]
- 19.Shen MD, Nordahl CW, Young GS, Wootton-Gorges SL, Lee A, Liston SE, et al. Early brain enlargement and elevated extra-axial fluid in infants who develop autism spectrum disorder. Brain. 2013;136:2825–2835. doi: 10.1093/brain/awt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carper RA, Moses P, Tigue ZD, Courchesne E. Cerebral lobes in autism: early hyperplasia and abnormal age effects. NeuroImage. 2002;16:1038–1051. doi: 10.1006/nimg.2002.1099. [DOI] [PubMed] [Google Scholar]
- 21.Sparks BF, Friedman SD, Shaw DW, Aylward EH, Echelard D, Artru AA, et al. Brain structural abnormalities in young children with autism spectrum disorder. Neurology. 2002;59:184–192. doi: 10.1212/wnl.59.2.184. [DOI] [PubMed] [Google Scholar]
- 22.Hazlett HC, Poe M, Gerig G, Smith RG, Provenzale J, Ross A, et al. Magnetic resonance imaging and head circumference study of brain size in autism: birth through age 2 years. Arch Gen Psychiatry. 2005;62:1366–1376. doi: 10.1001/archpsyc.62.12.1366. [DOI] [PubMed] [Google Scholar]
- 23.Kemper TL, Bauman M. Neuropathology of infantile autism. Journal of neuropathology and experimental neurology. 1998;57:645–652. doi: 10.1097/00005072-199807000-00001. [DOI] [PubMed] [Google Scholar]
- 24.Courchesne E, Mouton PR, Calhoun ME, Semendeferi K, Ahrens-Barbeau C, Hallet MJ, et al. Neuron number and size in prefrontal cortex of children with autism. JAMA. 2011;306:2001–2010. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- 25.Morgan JT, Chana G, Abramson I, Semendeferi K, Courchesne E, Everall IP. Abnormal microglial-neuronal spatial organization in the dorsolateral prefrontal cortex in autism. Brain research. 2012;1456:72–81. doi: 10.1016/j.brainres.2012.03.036. [DOI] [PubMed] [Google Scholar]
- 26.Hertz-Picciotto I, Delwiche L. The rise in autism and the role of age at diagnosis. Epidemiology. 2009;20:84–90. doi: 10.1097/EDE.0b013e3181902d15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King M, Bearman P. Diagnostic change and the increased prevalence of autism. Int J Epidemiol. 2009;38:1224–1234. doi: 10.1093/ije/dyp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers SM, Johnson CP, American Academy of Pediatrics Council on Children With D Management of children with autism spectrum disorders. Pediatrics. 2007;120:1162–1182. doi: 10.1542/peds.2007-2362. [DOI] [PubMed] [Google Scholar]
- 29.Warren Z, McPheeters ML, Sathe N, Foss-Feig JH, Glasser A, Veenstra-Vanderweele J. A systematic review of early intensive intervention for autism spectrum disorders. Pediatrics. 2011;127:e1303–1311. doi: 10.1542/peds.2011-0426. [DOI] [PubMed] [Google Scholar]
- 30.Buescher AV, Cidav Z, Knapp M, Mandell DS. Costs of autism spectrum disorders in the United Kingdom and the United States. JAMA pediatrics. 2014;168:721–728. doi: 10.1001/jamapediatrics.2014.210. [DOI] [PubMed] [Google Scholar]
- 31.Bruder MB, Kerins G, Mazzarella C, Sims J, Stein N. Brief Report: The Medical Care of Adults with Autism Spectrum Disorders: Identifying the Needs. J Autism Dev Disord. 2012 doi: 10.1007/s10803-012-1496-x. [DOI] [PubMed] [Google Scholar]
- 32.Kogan MD, Strickland BB, Blumberg SJ, Singh GK, Perrin JM, van Dyck PC. A national profile of the health care experiences and family impact of autism spectrum disorder among children in the United States, 2005-2006. Pediatrics. 2008;122:e1149–1158. doi: 10.1542/peds.2008-1057. [DOI] [PubMed] [Google Scholar]
- 33.Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC, Gao F, et al. Human-specific transcriptional networks in the brain. Neuron. 2012;75:601–617. doi: 10.1016/j.neuron.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuen RK, Thiruvahindrapuram B, Merico D, Walker S, Tammimies K, Hoang N, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 36.Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konopka G, Wexler E, Rosen E, Mukamel Z, Osborn GE, Chen L, et al. Modeling the functional genomics of autism using human neurons. Mol Psychiatry. 2012;17:202–214. doi: 10.1038/mp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stein JL, de la Torre-Ubieta L, Tian Y, Parikshak NN, Hernandez IA, Marchetto MC, et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron. 2014;83:69–86. doi: 10.1016/j.neuron.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geerts H. Of mice and men: bridging the translational disconnect in CNS drug discovery. CNS drugs. 2009;23:915–926. doi: 10.2165/11310890-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 40.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeFelipe J, Alonso-Nanclares L, Arellano JI. Microstructure of the neocortex: comparative aspects. J Neurocytol. 2002;31:299–316. doi: 10.1023/a:1024130211265. [DOI] [PubMed] [Google Scholar]
- 42.Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauman MD, Iosif AM, Ashwood P, Braunschweig D, Lee A, Schumann CM, et al. Maternal antibodies from mothers of children with autism alter brain growth and social behavior development in the rhesus monkey. Translational psychiatry. 2013;3:e278. doi: 10.1038/tp.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, et al. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell stem cell. 2014;14:323–328. doi: 10.1016/j.stem.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z, Zhou X, Zhu Y, Chen ZF, Yu B, Wang Y, et al. Generation of a monkey with MECP2 mutations by TALEN-based gene targeting. Neuroscience bulletin. 2014;30:381–386. doi: 10.1007/s12264-014-1434-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dragunow M. The adult human brain in preclinical drug development. Nat Rev Drug Discov. 2008;7:659–666. doi: 10.1038/nrd2617. [DOI] [PubMed] [Google Scholar]
- 47.Marchetto MC, Brennand KJ, Boyer LF, Gage FH. Induced pluripotent stem cells (iPSCs) and neurological disease modeling: progress and promises. Hum Mol Genet. 2011;20:R109–115. doi: 10.1093/hmg/ddr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 49.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 50.Freitas BC, Trujillo CA, Carromeu C, Yusupova M, Herai RH, Muotri AR. Stem cells and modeling of autism spectrum disorders. Exp Neurol. 2012 doi: 10.1016/j.expneurol.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han SS, Williams LA, Eggan KC. Constructing and deconstructing stem cell models of neurological disease. Neuron. 2011;70:626–644. doi: 10.1016/j.neuron.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 52.Beltrao-Braga PC, Pignatari GC, Russo FB, Fernandes IR, Muotri AR. In-a-dish: induced pluripotent stem cells as a novel model for human diseases. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2013;83:11–17. doi: 10.1002/cyto.a.22231. [DOI] [PubMed] [Google Scholar]
- 53.Kim JE, O'Sullivan ML, Sanchez CA, Hwang M, Israel MA, Brennand K, et al. Investigating synapse formation and function using human pluripotent stem cell-derived neurons. Proc Natl Acad Sci U S A. 2011;108:3005–3010. doi: 10.1073/pnas.1007753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Griesi-Oliveira K, Acab A, Gupta AR, Sunaga DY, Chailangkarn T, Nicol X, et al. Modeling non-syndromic autism and the impact of TRPC6 disruption in human neurons. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitne-Neto M, Machado-Costa M, Marchetto MC, Bengtson MH, Joazeiro CA, Tsuda H, et al. Downregulation of VAPB expression in motor neurons derived from induced pluripotent stem cells of ALS8 patients. Hum Mol Genet. 2011;20:3642–3652. doi: 10.1093/hmg/ddr284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y. Self-Organization of Polarized Cerebellar Tissue in 3D Culture of Human Pluripotent Stem Cells. Cell reports. 2015 doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 57.Cunningham M, Cho JH, Leung A, Savvidis G, Ahn S, Moon M, et al. hPSC-derived maturing GABAergic interneurons ameliorate seizures and abnormal behavior in epileptic mice. Cell stem cell. 2014;15:559–573. doi: 10.1016/j.stem.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim TG, Yao R, Monnell T, Cho JH, Vasudevan A, Koh A, et al. Efficient specification of interneurons from human pluripotent stem cells by dorsoventral and rostrocaudal modulation. Stem Cells. 2014;32:1789–1804. doi: 10.1002/stem.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen H, Qian K, Chen W, Hu B, Blackbourn LWt, Du Z, et al. Human-derived neural progenitors functionally replace astrocytes in adult mice. J Clin Invest. 2015 doi: 10.1172/JCI69097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeng X, Cai J, Chen J, Luo Y, You ZB, Fotter E, et al. Dopaminergic differentiation of human embryonic stem cells. Stem Cells. 2004;22:925–940. doi: 10.1634/stemcells.22-6-925. [DOI] [PubMed] [Google Scholar]
- 61.Krencik R, Zhang SC. Directed differentiation of functional astroglial subtypes from human pluripotent stem cells. Nature protocols. 2011;6:1710–1717. doi: 10.1038/nprot.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu GH, Suzuki K, Qu J, Sancho-Martinez I, Yi F, Li M, et al. Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell stem cell. 2011;8:688–694. doi: 10.1016/j.stem.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai X, Evrony GD, Lehmann HS, Elhosary PC, Mehta BK, Poduri A, et al. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell reports. 2014;8:1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cai M, Yang Y. Targeted genome editing tools for disease modeling and gene therapy. Current gene therapy. 2014;14:2–9. doi: 10.2174/156652321402140318165450. [DOI] [PubMed] [Google Scholar]
- 66.Cheung AY, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A, et al. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell stem cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rao M. iPSC crowdsourcing: a model for obtaining large panels of stem cell lines for screening. Cell stem cell. 2013;13:389–391. doi: 10.1016/j.stem.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 69.Muotri AR. Inducible pluripotent stem cells in autism spectrum disorders. In: Buxbaum JD, Hof naPR, editors. The Neuroscience of Autism Spectrum Disorders. 1st Elsevier; Oxford: 2013. pp. 391–400. [Google Scholar]
- 70.Belichenko PV, Oldfors A, Hagberg B, Dahlstrom A. Rett syndrome: 3-D confocal microscopy of cortical pyramidal dendrites and afferents. Neuroreport. 1994;5:1509–1513. [PubMed] [Google Scholar]
- 71.Tropea D, Giacometti E, Wilson NR, Beard C, McCurry C, Fu DD, et al. Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc Natl Acad Sci U S A. 2009;106:2029–2034. doi: 10.1073/pnas.0812394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ananiev G, Williams EC, Li H, Chang Q. Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from Rett syndrome patients as in vitro disease model. PLoS One. 2011;6:e25255. doi: 10.1371/journal.pone.0025255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim KY, Hysolli E, Park IH. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci U S A. 2011;108:14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 75.Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell stem cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 76.Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell stem cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PloS one. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doers ME, Musser MT, Nichol R, Berndt ER, Baker M, Gomez TM, et al. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem cells and development. 2014;23:1777–1787. doi: 10.1089/scd.2014.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 80.Krey JF, Pasca SP, Shcheglovitov A, Yazawa M, Schwemberger R, Rasmusson R, et al. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nature neuroscience. 2013;16:201–209. doi: 10.1038/nn.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chanda S, Marro S, Wernig M, Sudhof TC. Neurons generated by direct conversion of fibroblasts reproduce synaptic phenotype caused by autism-associated neuroligin-3 mutation. Proc Natl Acad Sci U S A. 2013;110:16622–16627. doi: 10.1073/pnas.1316240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 84.Shcheglovitov A, Shcheglovitova O, Yazawa M, Portmann T, Shu R, Sebastiano V, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503:267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schroer RJ, Phelan MC, Michaelis RC, Crawford EC, Skinner SA, Cuccaro M, et al. Autism and maternally derived aberrations of chromosome 15q. Am J Med Genet. 1998;76:327–336. doi: 10.1002/(sici)1096-8628(19980401)76:4<327::aid-ajmg8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 86.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 87.Germain ND, Chen PF, Plocik, AM, Glatt-Deeley H, Brown, J, Fink, JJ, Bolduc, KA, Robsinson, TM, Levine, ES, Reiter, LT, Graveley, BR, Lalande, M, Chamberlain, SJ. Gene expression analyses of human induced pluripotent stem cell-derived neurons carrying copy number variants of chromosome 15q11-q13.1. Molecular Autism. 2014;5:44–62. doi: 10.1186/2040-2392-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Adamo A, Atashpaz S, Germain PL, Zanella M, D'Agostino G, Albertin V, et al. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat Genet. 2015;47:132–141. doi: 10.1038/ng.3169. [DOI] [PubMed] [Google Scholar]
- 90.Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gutierrez RC, Hung J, Zhang Y, Kertesz AC, Espina FJ, Colicos MA. Altered synchrony and connectivity in neuronal networks expressing an autism-related mutation of neuroligin 3. Neuroscience. 2009;162:208–221. doi: 10.1016/j.neuroscience.2009.04.062. [DOI] [PubMed] [Google Scholar]
- 92.Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 93.Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.El-Fishawy P, State MW. The genetics of autism: key issues, recent findings, and clinical implications. Psychiatr Clin North Am. 2010;33:83–105. doi: 10.1016/j.psc.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Geschwind DH. Genetics of autism spectrum disorders. Trends Cogn Sci. 2011;15:409–416. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11. and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 97.Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS genetics. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.State MW, Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nat Neurosci. 2011 doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beltrao-Braga PI, Pignatari GC, Maiorka PC, Oliveira NA, Lizier NF, Wenceslau CV, et al. Feeder-free derivation of induced pluripotent stem cells from human immature dental pulp stem cells. Cell Transplant. 2011 doi: 10.3727/096368911X566235. [DOI] [PubMed] [Google Scholar]
- 100.Tai Y, Feng S, Ge R, Du W, Zhang X, He Z, et al. TRPC6 channels promote dendritic growth via the CaMKIV-CREB pathway. J Cell Sci. 2008;121:2301–2307. doi: 10.1242/jcs.026906. [DOI] [PubMed] [Google Scholar]
- 101.Zhou J, Du W, Zhou K, Tai Y, Yao H, Jia Y, et al. Critical role of TRPC6 channels in the formation of excitatory synapses. Nat Neurosci. 2008;11:741–743. doi: 10.1038/nn.2127. [DOI] [PubMed] [Google Scholar]
- 102.Greek R, Rice MJ. Animal models and conserved processes. Theoretical biology & medical modelling. 2012;9:40. doi: 10.1186/1742-4682-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van der Worp HB, Howells DW, Sena ES, Porritt MJ, Rewell S, O'Collins V, et al. Can animal models of disease reliably inform human studies? PLoS medicine. 2010;7:e1000245. doi: 10.1371/journal.pmed.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vierbuchen T, Wernig M. Direct lineage conversions: unnatural but useful? Nat Biotechnol. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Diniz LP, Almeida JC, Tortelli V, Vargas Lopes C, Setti-Perdigao P, Stipursky J, et al. Astrocyte-induced synaptogenesis is mediated by transforming growth factor beta signaling through modulation of D-serine levels in cerebral cortex neurons. The Journal of biological chemistry. 2012;287:41432–41445. doi: 10.1074/jbc.M112.380824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chung WS, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504:394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 110.Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15:477–486. doi: 10.1038/nn.3041. S471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nature protocols. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 113.Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SS, Sandoe J, et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell reports. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]