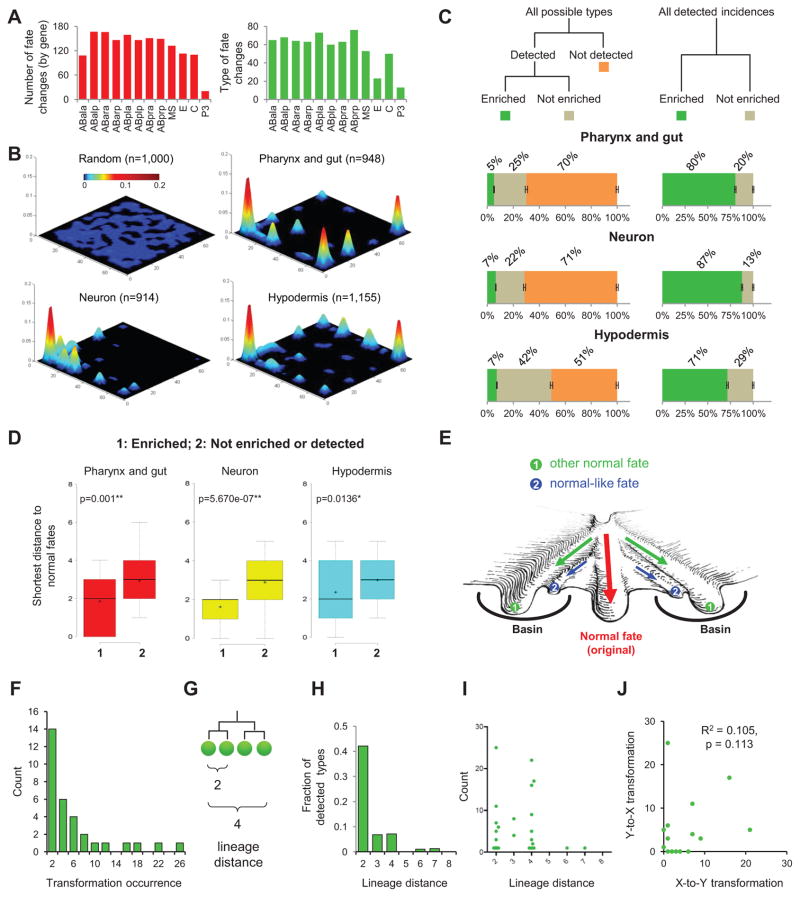
Figure 2. Canalized Landscape of Cell Fate.
(A) Summary of cell fate changes. Bar plots show the numbers (red) and types (green) of cell fate changes observed for the 12 founder cells.
(B) 3D plots show the phenotypic landscape of all detected progenitor cell fate changes. The X-Y plane is a phenotypic space with each type of cell fate being at a unique 2D coordinate. Z-axis shows the detected frequency of the fate. The first plot shows a random sampling from all 256 fate types. See also Figure S3.
(C) Statistics of cell fate phenotypes. Left panel shows the fraction of detected (enriched and not enriched) and not detected fates out of the 256 types for each tissue marker. Error bars show S.D. based on 10,000 simulations with known error rates of lineaging. Right panel shows the fraction of the enriched and not enriched types among the detected fate changes.
(D) Box plot shows the shortest distance of newly acquired fates to cell fates used in the wild type for different categories. Distance is quantified as the total number of clones whose marker expression status is different. In the box, horizontal lines and ‘+’ represent the median and mean respectively. P-value was calculated by two-tailed Mann-Whitney U Test. See also Figure S3.
(E) Artistic rendering of the canalization of differentiation around the fates used in normal development. Green arrows indicate homeotic transformations.
(F) Histogram of the observed occurrence of detected homeotic transformations.
(G) Definition of lineage distance.
(H) Fraction of detected types out of all possible types of homeotic transformations at different lineage distances.
(I) Scatter plot shows the number of occurrences and the lineage distance of each observed transformation.
(J) Scatter plot shows the correlation between the occurrence of X-to-Y and Y-to-X transformations for each detected fate pair.