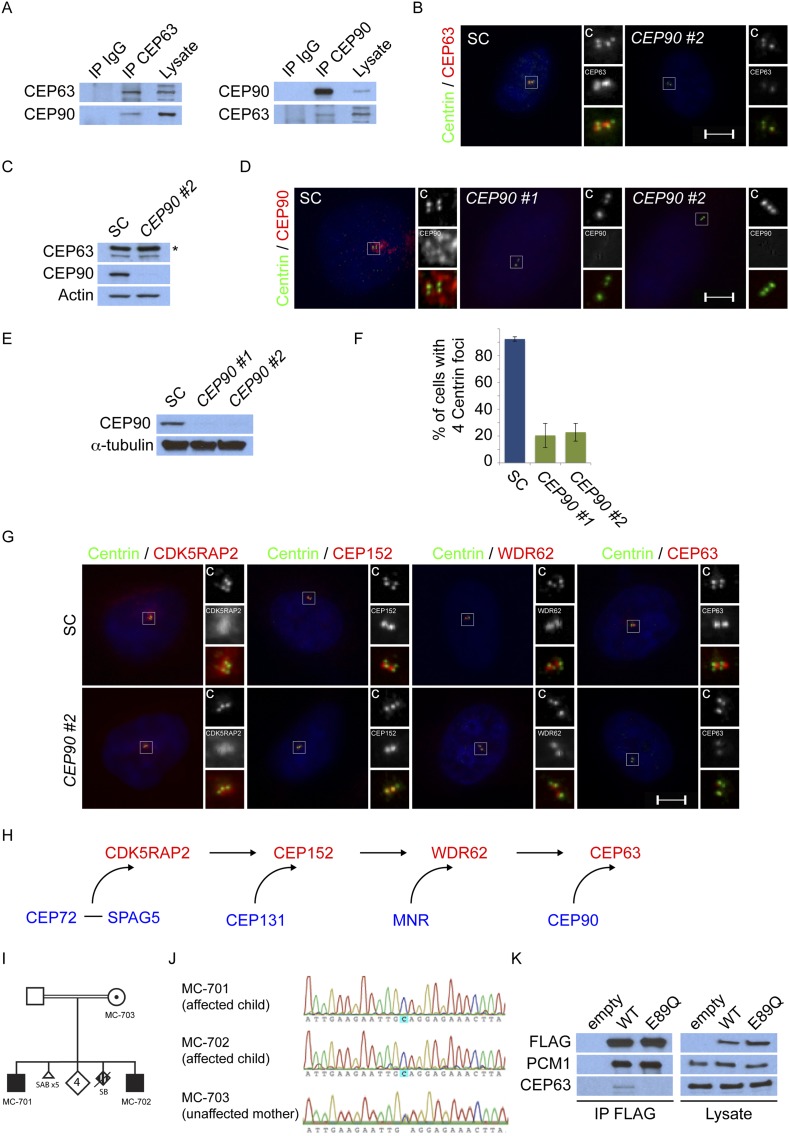
Figure 6. CEP90 encodes a centriolar satellite and MCPH-associated protein required to localize CEP63 to the centrosome to promote centriole duplication.
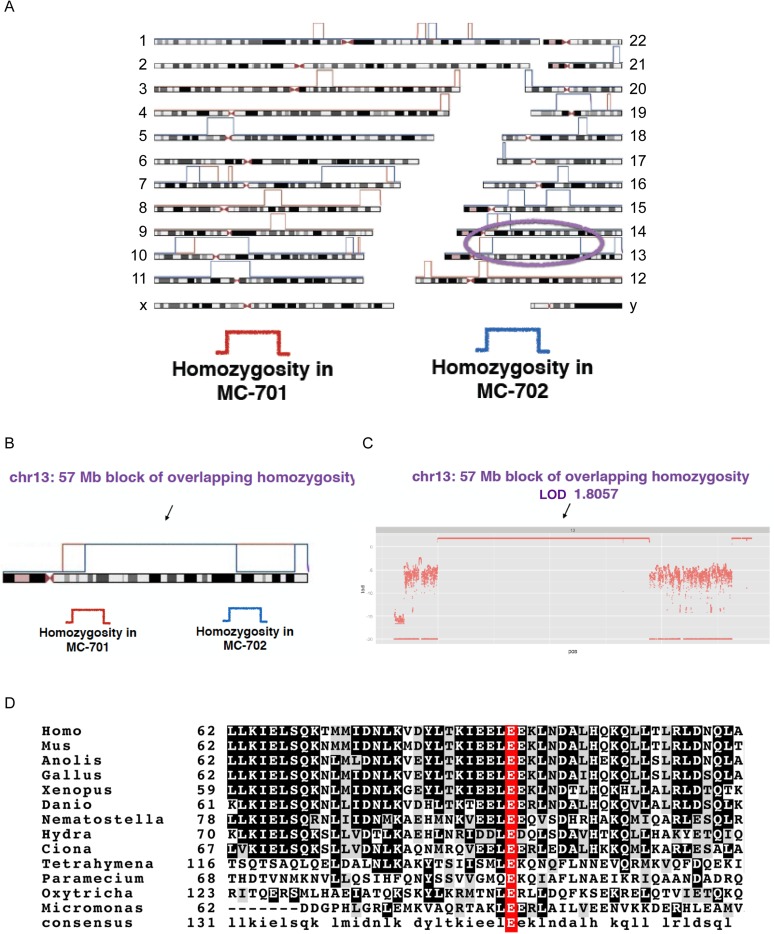
(A) HeLa total cell lysate was subjected to endogenous immunoprecipitation using antibodies to CEP63, CEP90 and a negative control. Precipitating proteins were analyzed by immunoblotting for CEP63 and CEP90. (B) Immunofluorescence images of SC and CEP90 #2 siRNA-treated HeLa cells co-stained for CEP63 (red) and Centrin (‘c’, green). (C) Total cell lysates of SC or CEP90 #2 siRNA transfected HeLa cells were analyzed by immunoblotting with antibodies to CEP63, CEP90 and Actin, which served as a loading control. Asterisk indicates the specific band. (D) S phase SC, CEP90 #1, and CEP90 #2 siRNA-treated cells were analyzed by immunofluorescence with antibodies to CEP90 (red) and Centrin (‘c’, green). (E) Total cell lysates of SC, CEP90 #1, and CEP90 #2 siRNA transfected HeLa cells were analyzed by immunoblotting for CEP90. α-tubulin served as a loading control. (F) Mean percentage of S phase SC, CEP90 #1, and CEP90 #2 siRNA-treated HeLa cells with four Centrin foci. At least 100 cells were counted per experiment (n = 3), p < 0.005 (paired t-test). (G) SC and CEP90 #2-depleted S phase cells were co-stained with Centrin (‘c’, green), CDK5RAP2 (red), CEP152 (red), WDR62 (red), and CEP63 (red). Scale bars indicate 5 μm for all images. (H) Schematic indicating that CEP90 is required for the centrosomal localization of CEP63. (I) Diagram of a simplified pedigree. Filled squares indicate individuals with MCPH. Spontaneous abortions (SAB), stillbirths (SB), and individuals of unknown gender (diamonds) are also indicated. (J) Sanger sequencing of the affected patients and their unaffected mother confirms the presence of a guanine to cytosine mutation leading to a charge reversing glutamic acid to glutamine substitution. This variant was identified by whole exome sequencing of the affected individuals and the unaffected mother. (K) We immunoprecipitated the FLAG tags of the wild-type and E89Q forms of CEP90 and blotted for endogenous CEP63 and FLAG.
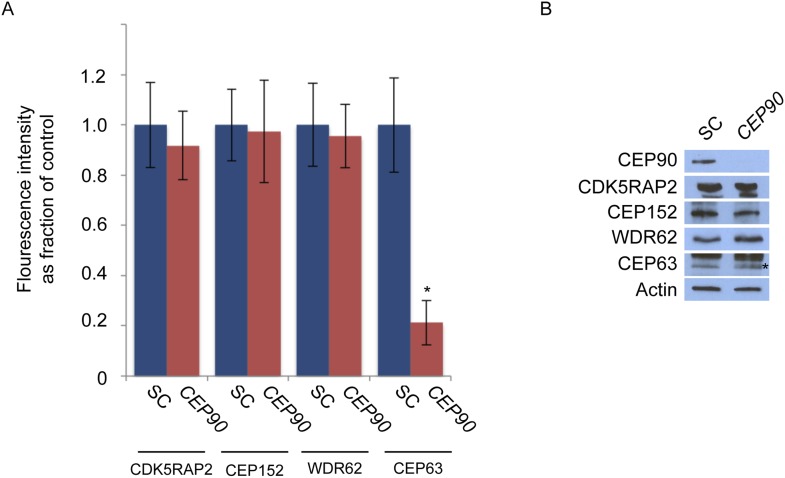
Figure 6—figure supplement 1. CEP90 is required to localize CEP63 to the centrosome.
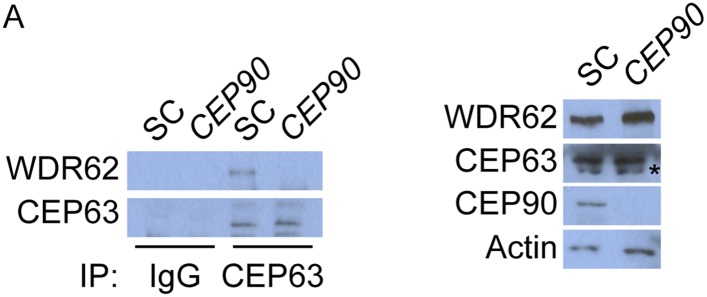
Figure 6—figure supplement 2. CEP63 interacts with WDR62 in a CEP90-dependent manner.