Figure 4.
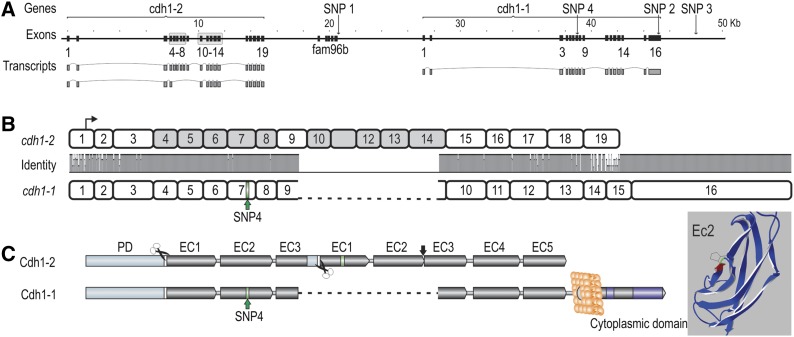
The core QTL region, containing cdh1-1, cdh1-2, and fam96b. (A) Organization of the cdh1 and fam96b genes in the core QTL region, with positions relative to ICSASG scaffold, ccf1000000016_0_0. Main transcripts of the cdh1 genes are shown. Gray boxes correspond to the duplicated exon blocks in cdh1-2. (B) Homology between full-length cdh1-1 and cdh1-2 transcripts. The former is encoded by 16 exons, whereas cdh1-2 consists of 19 exons. Transcriptional start is denoted by arrow and SNP4 by green bar on cdh1-1. Note the duplicated blocks of exons in cdh1-2 not present in cdh1-1. (C) The left figure shows the domain architecture of Cdh1-1 and Cdh1-2, where the former contains all epithelial cadherin domains of a fully functional protein, including a prodomain (orange), proteolytic cleavage signals (white bar/scissor), extracellular (EC) domains 1–5 (gray), transmembrane domain (light gray circle), and cytoplasmic tail with catenin binding motifs (purple). The protein residue in EC2 corresponding to SNP4 has been labeled in green. The full-length Cdh1-2 protein contains several anomalies when compared to a classical Cdh1. The duplicated exon blocks leads to duplications of EC1–EC3, though EC3 is truncated and fused to EC1, interspersed by a proteolytic cleavage site. EC2 and EC3 of the second exon block are fused and the residues C-terminal to EC5 are absent; hence, the protein is most likely not anchored to the cell membrane. The 3D structure of Cdh1-1 EC2 with the SNP4 S167P mutation is highlighted in green. The amino acid colored in red corresponds to the side of the hydrophobic pocket involved in cis-dimerization (Harrison et al. 2011).