Abstract
Two genes encoding protein components of the nuclear pore complex Nup160 and Nup96 cause lethality in F2-like hybrid genotypes between Drosophila simulans and Drosophila melanogaster. In particular, D. simulans Nup160 and Nup96 each cause inviability when hemizygous or homozygous in species hybrids that are also hemizygous (or homozygous) for the D. melanogaster X chromosome. The hybrid lethality of Nup160, however, is genetically complex, depending on one or more unknown additional factors in the autosomal background. Here we study the genetics and evolution of Nup160-mediated hybrid lethality in three ways. First, we test for variability in Nup160-mediated hybrid lethality within and among the three species of the D. simulans clade— D. simulans, D. sechellia, and D. mauritiana. We show that the hybrid lethality of Nup160 is fixed in D. simulans and D. sechellia but absent in D. mauritiana. Second, we explore how the hybrid lethality of Nup160 depends on other loci in the autosomal background. We find that D. simulans Nup160-mediated hybrid lethality does not depend on the presence of D. melanogaster Nup96, and we find that D. simulans and D. mauritiana are functionally differentiated at Nup160 as well as at other autosomal factor(s). Finally, we use population genetics data to show that Nup160 has experienced histories of recurrent positive selection both before and after the split of the three D. simulans clade species ∼240,000 years ago. Our genetic results suggest that a hybrid lethal Nup160 allele evolved before the split of the three D. simulans clade species, whereas the other autosomal factor(s) evolved more recently.
Keywords: speciation, hybrid incompatibility, hybrid inviability, nuclear pore complex
MANY species come to be reproductively isolated through the evolution of genetic incompatibilities that cause intrinsic sterility or inviability in interspecific hybrids (Dobzhansky 1937; Coyne and Orr 2004). Genetic substitutions that are neutral or favorable in the genetic background of one species can be severely deleterious when combined with the genetic background of another species (Dobzhansky 1937; Muller 1940, 1942). Decades of genetic analyses have provided broad support for this so-called Dobzhansky-Muller model—hybrid sterility and inviability typically result from incompatible epistatic interactions between two or more genetic factors (Coyne and Orr 2004). More recently, molecular analyses have turned to identifying the genes involved in hybrid incompatibilities and determining their evolutionary histories, functions within species, and dysfunctions in species hybrids. In Drosophila, these analyses have revealed that hybrid incompatibilities are typically genetically complex (involving three or more factors), targets of recurrent positive selection, and involved in genetic conflict with selfish genetic elements (Johnson 2010; Presgraves 2010a, b; Maheshwari and Barbash 2011).
For nearly a century, Drosophila melanogaster and Drosophila simulans have been important models for the study of hybrid incompatibilities owing to the genetic resources available in D. melanogaster (Sturtevant 1920; Provine 1991; Sawamura 2000; Barbash 2010). In crosses between D. melanogaster females and D. simulans males, the X-linked Hybrid male rescue (Hmr) gene of D. melanogaster is incompatible with the autosomal Lethal hybrid rescue (Lhr) gene of D. simulans, killing F1 hybrid males as late-stage larvae (Brideau et al. 2006). Both genes encode DNA-binding proteins that localize to centromeric heterochromatin (Thomae et al. 2013), affect expression of transposable elements and satellite DNA (Satyaki et al. 2014), and have histories of positive selection (Barbash et al. 2004; Brideau et al. 2006). The Hmr-Lhr hybrid incompatibility is genetically complex, requiring at least one additional unknown factor to cause hybrid lethality (Brideau et al. 2006). In the reciprocal cross, between D. simulans females and D. melanogaster males, the X-linked Zygotic hybrid rescue (Zhr) locus of D. melanogaster corresponds to a large species-specific pericentric block of 359-bp satellite DNA (Sawamura et al. 1995; Ferree and Barbash 2009) that is incompatible with an unidentified maternal factor, known as maternal hybrid rescue (mhr), present in many D. simulans lines (Sawamura et al. 1993; Orr 1996; Gerard and Presgraves 2012), killing F1 hybrid females as embryos (Hadorn 1961). Selfish repetitive DNA is implicated in the evolution of both Hmr-Lhr and mhr-Zhr hybrid incompatibilities.
At each of these loci, rescue mutations (compatible alleles) have been recovered that can rescue hybrids from lethality and, for some, hybrid female sterility (Watanabe 1979; Hutter and Ashburner 1987; Davis et al. 1996; Barbash and Ashburner 2003). These rescue mutations, when combined with other D. melanogaster tools (Sawamura et al. 2000; Presgraves 2003; Masly et al. 2006), have facilitated the mapping and identification of three additional hybrid incompatibility genes that affect F2-like hybrid genotypes. The male fertility-essential gene JYalpha has transposed from chromosome 4 in D. melanogaster to chromosome 3 in D. simulans, so hybrids homozygous for D. simulans chromosome 4 in an otherwise D. melanogaster genetic background completely lack JYalpha and are male sterile (Masly et al. 2006). And two D. simulans genes, Nup160 and Nup96, are incompatible with an unidentified factor(s) on the D. melanogaster X chromosome, causing hybrid lethality (Presgraves et al. 2003; Tang and Presgraves 2009; Sawamura et al. 2010).
Nup160 and Nup96 both encode protein components of the nuclear pore complex (NPC). The NPC mediates all molecular traffic between the cytoplasm and nucleus and interacts with DNA to regulate gene expression and chromatin organization (Capelson et al. 2010; Kalverda and Fornerod 2010; Liang and Hetzer 2011; Grossman et al. 2012). Its ∼30 different protein constituents (termed nucleoporins), protein-protein interactions, and overall architecture are largely conserved among eukaryotes (Bapteste et al. 2005; Neumann et al. 2010). Despite these deeply conserved functions, nucleoporins present some of the strongest evidence for recurrent adaptive protein evolution in the Drosophila genome (Begun et al. 2007; Presgraves and Stephan 2007; Langley et al. 2012; Nolte et al. 2013; Garrigan et al. 2014). Nup160 and Nup96 have histories of recurrent positive natural selection in both the D. melanogaster and D. simulans lineages (Presgraves et al. 2003; Tang and Presgraves 2009), leading to speculation that these and other nucleoporins have engaged in antagonistic co-evolutionary interactions with retroviruses, retrotransposons, or meiotic drive elements (Presgraves 2007; Presgraves and Stephan 2007). Nup160 is part of a complex genetic incompatibility, with hybrid lethality requiring the appropriate genotype at three or more loci. Hybrids die when homozygous or hemizygous for the D. melanogaster X chromosome (hereafter Xmel), homozygous or hemizygous for D. simulans Nup160 (hereafter Nup160sim), and heterozygous for the autosomes. Changing the genotype at any one of these loci—e.g., introducing a Nup160mel transgene, replacing Xmel with Xsim, or making the autosomal background homozygous D. melanogaster—suppresses hybrid lethality (Sawamura et al. 2004, 2010; Tang and Presgraves 2009).
In this report we further characterize the genetics and evolutionary history of the Nup160 hybrid incompatibility. First, we test for variability in Nup160-mediated hybrid lethality among the three species of the D. simulans clade—D. simulans, D. sechellia, and D. mauritiana—which diverged from one another ∼240,000 years ago (Kliman et al. 2000; Garrigan et al. 2012). We find that Nup160-mediated lethality occurs in hybrids from crosses of D. melanogaster with D. simulans and D. sechellia but not D. mauritiana. Interestingly, the same holds for Nup96 (Barbash 2007). Second, we test whether D. simulans and D. mauritiana have functionally diverged at Nup160 and/or at a different, genetically unlinked autosomal locus required to kill D. melanogaster hybrids. Third, because the NUP96 and NUP160 proteins physically interact at the NPC as part of the NUP107 subcomplex (Belgareh et al. 2001; Lutzmann et al. 2002), we test whether Nup160-mediated lethality depends on the hybrid genotype at Nup96 (and vice versa). Finally, we investigate the recent molecular evolutionary history of Nup160 among the three D. simulans clade species. Our genetic analyses suggest that a Nup160 allele with the capacity to cause hybrid lethality evolved before the split of the three D. simulans clade species but that only D. simulans and D. sechellia possess the additional autosomal factor(s) required for hybrid lethality.
Materials and Methods
Stocks, nomenclature, and crosses
The PiggyBac transposable element insertion PBac{RB}RfC38e00704 (obtained from the Exelixis Collection at Harvard Medical School) disrupts the overlapping 3′-untranslated regions of two genes, Nup160 and RfC38 (Thibault et al. 2004), and causes lethality in D. melanogaster–D. simulans hybrids that are hemizygous (XmelYsim) or homozygous for the D. melanogaster X chromosome (XmelXmelYsim). The Nup98E53.1 mutation disrupts the protein-coding sequence corresponding to the NUP96 protein and, similarly, causes lethality in D. melanogaster–D. simulans hybrids that are hemizygous (XmelYsim) or homozygous for the D. melanogaster X chromosome (XmelXmelYsim). For simplicity, we hereafter refer to PBac{RB}RfC38e00704 as Nup160− and Nup98E53.1 as Nup96−. To rescue F1 hybrid males resulting from crosses between D. melanogaster females and D. simulans, D. sechellia, and D. mauritiana males, we used the y w Df(1)Hmr v chromosome (provided by D. Barbash, Cornell University), which bears a partial deletion of Hybrid male rescue (Hmr) that removes the first 96 amino acids of the HMR protein (Barbash and Lorigan 2007). To rescue hybrid males in a different cross, we used D. simulans w; Lhr1, which has a mutation in the Lethal hybrid rescue (Lhr) gene. To test for variation in Nup160-mediated hybrid lethality within and among the D. simulans clade species, we used 10 D. simulans isofemale lines collected from Zimbabwe (provided by C. Aquadro, Cornell University), 6 D. sechellia isofemale lines (provided by J. Coyne, University of Chicago, and from the Tucson Stock Center), and 8 D. mauritiana isofemale lines (provided by J. Coyne, University of Chicago, and M. Ramos Womack, Princeton University).
To introgress Nup160sim from D. simulans into D. mauritiana white (w), we used a D. simulans strain with a tightly linked PiggyBac transgene bearing the visible dominant marker eYFP inserted on chromosome arm 2L (provided by D. Stern, Janelia Farm Research Campus, Howard Hughes Medical Institute). This PBac{eYFP} insert is at genomic sequence coordinate 2L:11,756,533 (D. Stern, personal communication), and Nup160 is at 2L:11,123,777–11,129,335 (on the D. melanogaster reference assembly version R6.02), placing the YFP marker ∼627 kb proximal to Nup160. We crossed mau w females to sim YFP males and collected fertile hybrid sim YFP/mau females; we then backcrossed sim YFP/mau virgin females to mau w males recurrently for six generations. After generation six, the sim YFP introgression was maintained by selection through males (without further recombination). To make the complementary introgression of Nup160mau from D. mauritiana into D. simulans w501, we used two D. mauritiana strains that each have a P-element transgene bearing the dominant visible mini-w+ marker on chromosome arm 2L (True et al. 1996). These insertions, P{w+}4G4C and P{w+}4G5, are located at coordinates 2L:12,700,666 and 2L:12,700,661(Araripe et al. 2006), placing them both ∼1.57 Mb proximal to Nup160. Briefly, we crossed sim w501 females to mau P{w+} males and collected fertile hybrid mau P{w+}/sim females; we then backcrossed mau P{w+}/sim virgin females to sim w501 males recurrently for six generations. After generation six, the mau P{w+} introgressions were maintained by selection through males (without further recombination). We confirmed the presence of Nup160sim and Nup160mau alleles in the introgression lines by sequencing a 550-bp PCR amplicon from exon 6 of Nup160 (details later).
All crosses were done at 22–23°. For each cross, at least two replicates were set up with ∼20 virgin females and ∼25 males. After larvae appeared, parents were transferred to fresh vials every 3 days for as long as they continued to produce progeny. Hybrid progeny were scored for at least 14 days after eclosion of the first fly until no more flies eclosed for 3 consecutive days.
Lethal phase
To determine the lethal phase of Nup160-mediated lethality, we crossed D. melanogaster y w; Nup160−/CyO females to D. simulans Lhr males. Hybrid male larvae from this cross have yellow mouthparts, whereas hybrid female larvae have black (wild-type) mouthparts. We collected hybrid larvae at the third instar stage, separated them by sex, transferred them to vials with fresh food, and then, following pupation, checked whether the pupae were alive or dead every ∼12 hr.
Sequencing of Nup160 and molecular population genetics analysis
To sequence Nup160 from D. mauritiana and D. sechellia lines, we first extracted genomic DNA from single females using the PUREGENE DNA Purification Kit (Gentra Systems, Minneapolis, MN). The entire ∼5.1-kb coding region of the Nup160 gene was PCR amplified with the Expand Long Template PCR System (Roche Applied Science, Indianapolis, IN). Both strands of the PCR products were sequenced directly with internal primers with ABI Prism BigDye Terminator chemistry (Applied Biosystems, Foster City, CA) on an automated ABI sequencer at the University of Rochester Functional Genomics Core Facility. All sequences were assembled and manually checked using Sequencher v. 4.5 (Genecodes, Ann Arbor, MI) and then manually aligned in Se-Al v. 2.0 (http://tree.bio.ed.ac.uk/software/seal/). Sequences have been deposited in GenBank under accession numbers KR817621–KR817638. Nup160 sequences from D. melanogaster and D. simulans are from Tang and Presgraves (2009) (GenBank accession numbers FJ600378–FJ600401). All population genetics analyses were performed with DnaSP v.5 (Librado and Rozas 2009).
Results
Effect of Nup160 in hybrids between D. melanogaster and the three D. simulans clade species
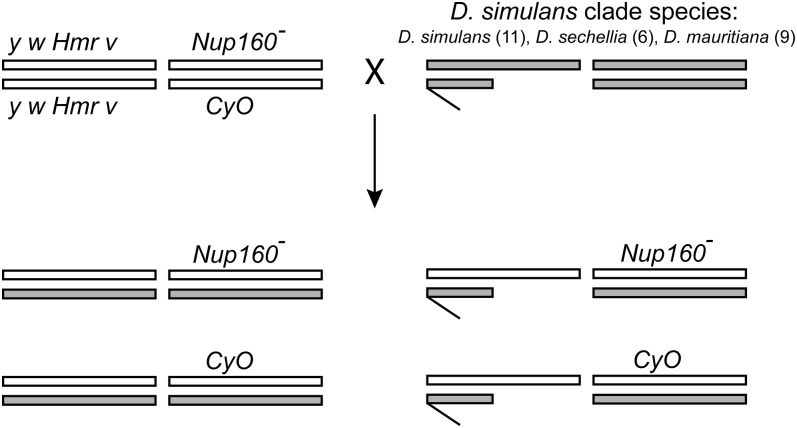
We first tested whether Nup160-dependent lethality is fixed in D. simulans. We crossed D. melanogaster y w Df(1)Hmr v; Nup160−/CyO females to D. simulans (sim) males from 11 isofemale lines (Figure 1). The resulting Nup160− hybrids must develop using only wild-type D. simulans Nup160. We found that the viability of the resulting Nup160−/sim hybrid females was lower than that of CyO/sim hybrid females in all but one cross (sim 036) and significantly lower in four crosses (Table 1). Overall, the relative viability of Nup160−/sim hybrid females is ∼75% that of CyO/sim hybrid females. While we recovered abundant CyO/sim hybrid males, we failed to recover any Nup160−/sim hybrid males (Table 1). These results confirm that Nup160-mediated lethality occurs in Hmr-bearing hybrid males and is not therefore a special property of the D. simulans Lhr1 genotypes used in previous analyses (Tang and Presgraves 2009; Sawamura et al. 2010). These results also show that Nup160-dependent hybrid lethality appears to be fixed in D. simulans (Table 1).
Figure 1.
Testing for Nup160-mediated lethality in hybrid males between D. melanogaster and the three D. simulans clade species. D. melanogaster y w Df(1)Hmr v; Nup160−/CyO females were crossed to males from multiple isofemale lines of D. simulans (n = 11), D. sechellia (n = 6), and D. mauritiana (n = 9). For each, sex chromosome and second chromosome genotypes are shown (third and fourth chromosomes not shown), with D. melanogaster = white and D. simulans, D. sechellia, or D. mauritiana = gray.
Table 1. Nup160-mediated hybrid lethality is fixed in D. simulans and D. sechellia but absent in D. mauritiana.
| Hybrid females | Hybrid males | ||||||
|---|---|---|---|---|---|---|---|
| Male parenta | Nup160− | CyO | Relative viability | Nup160− | CyO | Relative viability | Percent rescue |
| D. simulans | |||||||
| sim 005 | 24 | 47 | 0.51** | 0 | 20 | 0.00** | 42.6 |
| sim 007 | 32 | 50 | 0.64* | 0 | 21 | 0.00** | 42.0 |
| sun 017 | 34 | 35 | 0.97 | 0 | 15 | 0.00** | 42.9 |
| sim 019 | 50 | 59 | 0.85 | 0 | 18 | 0.00** | 30.5 |
| sim 026 | 34 | 44 | 0.77 | 0 | 14 | 0.00** | 31.8 |
| sim 036 | 59 | 49 | 1.20 | 0 | 21 | 0.00** | 42.9 |
| sim 049 | 14 | 27 | 0.52* | 0 | 14 | 0.00** | 51.9 |
| sim 058 | 21 | 33 | 0.64 | 0 | 16 | 0.00** | 48.5 |
| sim 065 | 37 | 46 | 0.80 | 0 | 20 | 0.00** | 43.5 |
| sim 224 | 48 | 77 | 0.62** | 0 | 32 | 0.00** | 41.6 |
| sim w501 | 69 | 83 | 0.83 | 0 | 27 | 0.00** | 32.5 |
| Total | 422 | 550 | 0.77** | 0 | 218 | 0.00** | 39.6 |
| D. sechellia | |||||||
| sec Robertson | 52 | 65 | 0.80 | 3 | 31 | 0.10** | 47.7 |
| sec Praslin | 44 | 56 | 0.79 | 0 | 18 | 0.00** | 32.1 |
| sec w | 39 | 33 | 1.18 | 1 | 15 | 0.07** | 45.5 |
| sec sy007 | 45 | 57 | 0.79 | 2 | 33 | 0.06** | 57.9 |
| sec sy034 | 46 | 88 | 0.52** | 1 | 30 | 0.03** | 34.1 |
| sec iso81 | 33 | 63 | 0.52** | 1 | 16 | 0.06** | 25.4 |
| Total | 259 | 362 | 0.72** | 8 | 143 | 0.06** | 39.5 |
| D. mauritiana | |||||||
| mau iso105 | 49 | 50 | 0.98 | 17 | 15 | 1.13 | 30.0 |
| mau iso152 | 58 | 62 | 0.94 | 30 | 24 | 1.25 | 38.7 |
| mau R11 | 70 | 53 | 1.32 | 22 | 16 | 1.38 | 30.2 |
| mau R23 | 41 | 36 | 1.14 | 8 | 19 | 0.42* | 52.8 |
| mau R35 | 39 | 43 | 0.91 | 13 | 19 | 0.68 | 44.2 |
| mau R45 | 47 | 45 | 1.04 | 17 | 11 | 1.55 | 24.4 |
| mau R48 | 53 | 49 | 1.08 | 23 | 18 | 1.28 | 36.7 |
| mau R57 | 30 | 30 | 1.00 | 13 | 16 | 0.81 | 53.3 |
| mau w | 215 | 205 | 1.05 | 82 | 73 | 1.12 | 35.6 |
| Total | 602 | 573 | 1.05 | 225 | 211 | 1.07 | 36.8 |
P < 0.05, **P < 0.01, χ2-tests.
For all crosses, D. melanogaster y w Df(1)Hmr v; Nup160−/CyO females were crossed to D. simulans, D. sechellia, or D. mauritiana males.
To determine the lethal phase of the Nup160 incompatibility, we crossed D. melanogaster y w; Nup160−/CyO females to D. simulans w; Lhr1 males, collected and separated hybrid male and female larvae based on the color of the larval mouthparts, and then scored viability of pupae every 12 hr. At 60–72 hr after pupa formation, about half the male pupae (12 of 29) were dead, as evidenced by deterioration of morphological structures and darkening of tissue (Table 2). All remaining 17 males eclosed at ∼132 hr after pupa formation, and all were CyO/sim. Among hybrid females, 34 of 36 eclosed at ∼120 hr after pupa formation, and 15 were CyO/sim. We checked the two dead female pupae by opening the pupal cases and found that unlike the dead male pupae, they were fully developed but failed to eclose (their wing phenotypes could not be scored). These findings show that Nup160-mediated hybrid lethality occurs at an early pupal stage [consistent with the findings of Maehara et al. (2012)], prior to the establishment of adult morphological structures. Nup160-mediated lethality thus occurs later than Hmr-Lhr-mediated hybrid lethality (late-stage larvae) (Sturtevant 1920) but earlier than Nup96-mediated hybrid lethality (late-stage pupae to pharate adult) (Barbash 2007).
Table 2. XmelYsim; Nup160−/sim hybrid males die as pupae at ∼60 hr of development.
| Hybrid males (n = 29) | Hybrid females (n = 36) | |||
|---|---|---|---|---|
| Time (hrs)a | Dead | Eclosed | Dead | Eclosed |
| 12–48 | 0 | 0 | 0 | 0 |
| 60 | 11 | 0 | 0 | 0 |
| 72 | 1 | 0 | 0 | 0 |
| 84–108 | 0 | 0 | 0 | 0 |
| 120 | 0 | 9 | 0 | 34 |
| 132 | 0 | 8 | 2 | 0 |
| Totals | 12 | 17 | 2 | 34 |
Hybrid third instar larvae were collected, separated by sex, and checked every 12 hr for lethality (see Materials and Methods).
We next tested whether Nup160-mediated hybrid lethality occurs in D. melanogaster–D. sechellia hybrids and D. melanogaster–D. mauritiana hybrids. We crossed D. melanogaster y w Df(1)Hmr v; Nup160−/CyO females to males from six D. sechellia isofemale lines and nine D. mauritiana isofemale lines. Nup160−/sech hybrid females show reduced relative viability in five of six crosses, two significantly so (∼72%) (Table 1), and Nup160−/sech hybrid males are effectively lethal (relative viability ∼6%) (Table 1). These results show that Nup160-mediated hybrid lethality is fixed in D. sechellia and behaves similarly to D. simulans, with moderate effects in hybrid females and lethal effects (or nearly so) in hybrid males. In contrast, Nup160−/mau hybrid females and Nup160−/mau hybrid males show no reduced viability relative to control CyO/mau siblings (Table 1). Interestingly, the effects of Nup160— lethal in XmelYsim and XmelYsech hybrids but not in XmelYmau hybrids—parallels that observed for Nup96 (Barbash 2007). These results together raised the possibility that the Nup160 and Nup96 hybrid incompatibilities may not be genetically independent.
The hybrid lethal effect of Nup160sim does not require the presence of Nup96mel
Given that the NUP96 and NUP160 proteins are predicted to physically interact with one another at the NPC (Belgareh et al. 2001; Lutzmann et al. 2002), we tested the possibility that the lethal effect of Nup160sim in hybrid males specifically requires the presence of Nup96mel. We constructed D. melanogaster Nup160−/CyO; Nup96−/TM3, Ser double-mutant females and crossed them to D. simulans w; Lhr1 males. This cross produces four hybrid female genotypes and four hybrid male genotypes. All four hybrid female genotypes are viable, occurring in expected ratios (Table 3). And, as expected, hybrid males with both the Nup160−/sim; TM3, Ser/sim and CyO/sim; Nup96−/sim genotypes are inviable (Table 3) (Presgraves et al. 2003; Tang and Presgraves 2009; Sawamura et al. 2010; Maehara et al. 2012). If Nup160sim requires Nup96mel to cause hybrid lethality (or vice versa), then double-mutant Nup160−/sim; Nup96−/sim hybrid males ought to be viable. Instead, we failed to recover any hybrid males with the genotype Nup160−/sim; Nup96−/sim. We conclude that Nup160sim does not require Nup96mel (and, similarly, that Nup96sim does not require Nup160mel) to kill hybrids. These findings are consistent with two possibilities: Nup160sim and Nup96sim behave as loss-of-function alleles in the hybrid genetic background or Nup160sim and Nup96sim have neomorphic lethal effects in hybrids, but these do not require the presence of Nup160mel and/or Nup96mel.
Table 3. Nup160-mediated hybrid lethality does not depend on Nup96 genotype, and vice versa.
| Autosomal genotype | |||||
|---|---|---|---|---|---|
| CyO/sim; Nup96−/sim | CyO/sim; TM3,Ser/sim | Nup160−/sim; Nup96−/sim | Nup160−/sim; TM3,Ser/sim | ||
| Replicate | Hybrid sex | ||||
| 1 | Xmel/Ysim | 0 | 29 | 0 | 0 |
| Xmel/Xsim | 31 | 40 | 50 | 40 | |
| 2 | Xmel/Ysim | 1 | 32 | 0 | 0 |
| Xmel/Xsim | 48 | 44 | 57 | 58 | |
| 3 | Xmel/Ysim | 0 | 18 | 0 | 0 |
| Xmel/Xsim | 31 | 47 | 56 | 43 | |
| Total | Xmel/Ysim | 1 | 79 | 0 | 0 |
| Xmel/Xsim | 110 | 131 | 163 | 141 | |
Lineage-specific Nup160-mediated hybrid lethality depends on Nup160 and autosomal background
Previous work established that the Nup160 hybrid incompatibility is complex, requiring (1) hemizygosity (or homozygosity) for Xmel, (2) hemizygosity (or homozygosity) for Nup160sim, and (3) at least one unknown dominant factor in the D. simulans autosomes (Sawamura et al. 2004, 2010, 2014). Our finding that Nup160-mediated lethality occurs in D. simulans–D. melanogaster hybrid males but not in D. mauritiana–D. melanogaster hybrid males therefore raises two nonexclusive possibilities: D. simulans and D. mauritiana are functionally differentiated at Nup160 and/or at some other autosomal factor(s) essential for hybrid lethality.
To test these possibilities, we performed reciprocal introgression experiments. First, we introgressed Nup160mau (marked with a tightly linked dominant visible marker, P{w+}; see Materials and Methods) into an otherwise D. simulans genetic background, backcrossing through fertile hybrid females for six generations. We then crossed D. melanogaster y w Df(1)Hmr v; Nup160−/CyO females to D. simulans males heterozygous for the introgression sim w501/Y; Nup160mau P{w+}/Nup160sim. This cross yields four hybrid female genotypes and four hybrid male genotypes: non-P{w+} hybrids are Nup160−/Nup160sim and CyO/Nup160sim, and P{w+} hybrids are Nup160−/Nup160mau and CyO/Nup160mau. As Table 4 shows, Nup160−/Nup160mau and Nup160−/Nup160sim hybrid male genotypes are both inviable. This finding shows that Nup160mau causes complete hybrid lethality when combined with at least one other dominant autosomal factor from D. simulans. Because Nup160mau does not cause hybrid lethality when on its own autosomal background, we conclude that at least one autosomal background factor required for hybrid lethality has functionally diverged between D. mauritiana and D. simulans.
Table 4. Nup160-mediated hybrid lethality depends on species-specific allele and genetic background.
| Hybrid females | Hybrid males | Percent rescueb | |||||
|---|---|---|---|---|---|---|---|
| Nup160− | CyO | Relative viability | Nup160− | CyO | Relative viability | ||
| Nup160mau P{w+}4G4C | 46 | 47 | 0.98 | 0 | 27 | 0.00* | 57.4 |
| Nup160sim | 54 | 40 | 1.35 | 0 | 22 | 0.00* | 55.0 |
| Nup160mau P{w+}4G5 | 111 | 68 | 1.63* | 0 | 68 | 0.00* | 100.0 |
| Nup160sim | 124 | 92 | 1.35 | 0 | 82 | 0.00* | 89.1 |
| Nup160sim.YFP | 282 | 271 | 1.04 | 19 | 76 | 0.25* | 28.0 |
| Nup160mau | 325 | 297 | 1.09 | 155 | 142 | 1.09 | 47.8 |
Alternative second chromosomes transmitted by Nup160 introgression males crossed to D. melanogaster Hmr; Nup160−/CyO females.
Percent rescue = (number of CyO hybrid males/number of CyO hybrid females) × 100.
P<0.05, χ2-test.
Next, we introgressed Nup160sim (marked with a tightly linked dominant visible marker, PBac{eYFP}; see Materials and Methods) into a largely D. mauritiana genetic background, backcrossing through fertile hybrid females for six generations. We then crossed D. melanogaster y w Df(1)Hmr v; Nup160−/CyO females to D. mauritiana males heterozygous for the introgression mau w/Y; Nup160sim YFP/Nup160mau. This cross yields four hybrid female genotypes and four hybrid male genotypes: non-YFP hybrids are Nup160−/Nup160mau and CyO/Nup160mau, and YFP hybrids are Nup160−/Nup160sim and CyO/Nup160sim. As Table 4 shows, Nup160−/Nup160mau (non-YFP) hybrid males are viable, but Nup160−/Nup160sim (YFP) hybrid males are sublethal (relative viability ∼25%). These findings show that the Nup160 alleles of the two species are functionally divergent: Nup160sim causes partial lethality in a D. mauritiana autosomal background, but Nup160mau does not. The fact that Nup160sim causes incomplete hybrid lethality in a D. mauritiana autosomal background (as opposed to the complete lethality observed in a D. simulans autosomal background) (Table 1) further suggests that the autosomal background factor(s) also has functionally diverged between D. simulans and D. mauritiana. However, we cannot exclude the possibility that an unknown autosomal factor, tightly linked to the Nup160 locus, was co-introgressed with Nup160sim, facilitating its hybrid lethal effect.
Molecular evolution of Nup160 in the D. simulans clade
Since the split of D. melanogaster and D. simulans, both lineages have experienced recurrent positive selection at Nup160 (Table 5, line 1) (Tang and Presgraves 2009). Here we extend these analyses by surveying polymorphism and divergence at Nup160 (∼5.1 kb) from 8 D. sechellia and 10 D. mauritiana lines. McDonald-Kreitman (MK) tests (McDonald and Kreitman 1991) reject the neutral hypothesis because Nup160 has an excess of fixed nonsynonymous differences among all three pairs of D. simulans clade species in pooled analyses (Table 5, lines 2–4). With D. yakuba Nup160 as a distant outgroup species, we parsimony-mapped nonsynonymous and synonymous substitutions onto five branches of the species phylogeny: the D. melanogaster terminal branch, the D. simulans clade ancestor internal branch, the D. simulans terminal branch, the D. sechellia terminal branch, and the D. mauritiana terminal branch. Codons experiencing multiple substitutions over the five-species history were excluded from the branch-specific analyses because they cannot be mapped unambiguously to particular branches. Using mapped substitutions, we asked whether recurrent adaptive evolution occurred in all lineages or in a subset. Furthermore, by mapping substitutions to the internal branch of the D. simulans clade ancestor, we asked whether Nup160 experienced positive selection before, after, or before and after the split of the three D. simulans clade species ∼240,000 years ago (Garrigan et al. 2012). To perform the MK test for the internal branch, we assumed that the population of the D. simulans clade ancestor had the same numbers of nonsynonymous and synonymous polymorphisms as found in the extant D. simulans population (Table 5, line 6). These branch-specific MK tests provide strong evidence for recurrent adaptive evolution at Nup160 in the common ancestor of the D. simulans clade (prior to 240,000 years ago) as well as within D. simulans (since ∼240,000 years ago) (Table 5, lines 6 and 7). The MK tests for D. mauritiana and D. sechellia lineages did not reject the neutral null hypothesis (Table 5, lines 8 and 9). None of the three D. simulans clade species showed evidence of a recent and/or strong selective sweep: mean silent nucleotide diversity at Nup160 is comparable to that of other autosomal loci in these species (Kliman et al. 2000; Legrand et al. 2011), and neither Tajima’s D (Tajima 1989) nor Fay and Wu’s H (Fay and Wu 2000), two summaries of the site-frequency spectra, revealed significant deviations from standard neutral expectations in any of the three species (Table 6).
Table 5. Evidence for lineage-specific recurrent adaptive protein evolution at Nup160.
| Polymorphic | Divergent | Fisher’s exact P-value | ||||||
|---|---|---|---|---|---|---|---|---|
| Line | R | S | R/S | R | S | R/S | ||
| 1 | D. melanogaster–D. simulans pooleda | 27 | 154 | 0.175 | 58 | 64 | 0.906 | 9.3 × 10−10 |
| 2 | D. simulans–D.mauritiana pooled | 34 | 168 | 0.202 | 20 | 4 | 5.000 | 8.2 × 10−11 |
| 3 | D. simulans–D. sechellia pooled | 19 | 107 | 0.178 | 27 | 20 | 1.350 | 1.1 × 10−7 |
| 4 | D. sechellia–D. mauritiana pooled | 19 | 87 | 0.218 | 12 | 18 | 0.667 | 0.015 |
| 5 | D. melanogaster lineage | 10 | 56 | 0.179 | 18 | 32 | 0.563 | 0.015 |
| 6 | D. simulans clade ancestral lineage | 17b | 100b | 0.170 | 24 | 15 | 1.600 | 4.4 × 10−8 |
| 7 | D. simulans lineage | 17 | 100 | 0.170 | 12 | 3 | 4.000 | 4.4 × 10−7 |
| 8 | D. sechellia lineage | 2 | 7 | 0.286 | 8 | 6 | 1.333 | 0.197 |
| 9 | D. mauritiana lineage | 34 | 168 | 0.202 | 1 | 0 | — | 0.172 |
D. melanogaster and D. simulans data are from Tang and Presgraves (2009).
For this MK test, the D. simulans clade ancestral population is assumed to have the same numbers of nonsynonymous and synonymous polymorphisms as the extant D. simulans population sample.
Table 6. Summaries of DNA sequence polymorphism at Nup160 in four Drosophila species.
| Species | na | bpb | Sc | πd | πsilente | Tajima’s D | FWHf |
|---|---|---|---|---|---|---|---|
| D. melanogaster | 12 | 5037 | 96 | 0.0059 | 0.0151 | −0.315 | −0.839 |
| D. simulans | 12 | 5009 | 188 | 0.0128 | 0.0328 | 0.102 | 0.192 |
| D. sechellia | 8 | 5037 | 8 | 0.0007 | 0.0016 | −0.312 | 0.432 |
| D. mauritiana | 10 | 5024 | 151 | 0.0095 | 0.0244 | −0.595 | 0.221 |
n = number of chromosomes sampled from each species.
bp = number of nucleotides in the intraspecies alignment.
S = the number of segregating sites.
π = average nucleotide diversity at all sites.
πsilent = average nucleotide diversity at silent sites.
FWH = normalized Fay & Wu’s H.
Discussion
Our work reveals two main findings. The first is that Nup160-mediated lethality in hybrids between D. melanogaster and its sibling species is fixed in D. simulans (n = 11) and in D. sechellia (n = 6) but absent from D. mauritiana (n = 9). Previous work established that the lethal hybrid incompatibility between Nup160sim and Xmel requires at least one additional (unknown) dominant autosomal factor from D. simulans (Sawamura et al. 2004, 2010). Consistent with this result, our second main finding is that Nup160 and at least one additional autosomal factor required for hybrid lethality are functionally divergent between D. simulans and D. mauritiana: Nup160mau kills hybrids when in a D. simulans autosomal background but not its own (Tables 1 and 4), showing that the D. simulans and D. mauritiana autosomal backgrounds are functionally different, and in a D. mauritiana autosomal background, Nup160sim causes partial hybrid lethality, but Nup160mau does not (Tables 1 and 4), showing that the two Nup160 alleles are functionally different.
Our genetic analyses allow several inferences about the phylogenetic history of the Nup160 hybrid incompatibility. For Nup160, the alleles of all three D. simulans clade species can cause hybrid lethality (Nup160sim and Nup160sech on their respective autosomal backgrounds and Nup160mau when introgressed into a D. simulans autosomal background). Therefore, the capacity of Nup160 to cause hybrid lethality almost certainly evolved in the common ancestor of the D. simulans clade species. For the autosomal background factor(s), our finding that the Nup160 hybrid incompatibility kills hybrids from crosses with D. simulans and D. sechellia but not D. mauritiana raises three possibilities. First, the autosomal factor(s) evolved in the common ancestor of all three species but was reversed subsequently in D. mauritiana. This scenario seems doubtful, requiring the incidental chance reversal of hybrid incompatibility (there is no selection favoring compatibility of Nup160mau with D. melanogaster). Second, the autosomal factor(s) is shared in D. simulans and D. sechellia owing to common ancestry—it either evolved in the common ancestor of these two species after the split from D. mauritiana or it evolved in one species and was exported to the other via gene flow (Garrigan et al. 2012; Matute and Ayroles 2014). Third, the autosomal factor(s) in D. simulans and D. sechellia may have converged independently on hybrid lethality. The second and third scenarios both imply that the necessary components of the complex Nup160 hybrid incompatibility evolved more recently than ∼240,000 years ago.
Like Nup160, Nup96-mediated hybrid lethality is lineage specific, genetically complex, and likely of relatively recent origin. Nup96sim and Nup96sech cause hybrid lethality when combined with hemizygous (or homozygous) Xmel, but Nup96mau does not (Presgraves et al. 2003; Barbash 2007). However, the fact that Nup96 in D. simulans has experienced no nonsynonymous substitutions since its split from D. mauritiana implies that Nup96sim and Nup96mau are functionally equivalent (Presgraves et al. 2003). Therefore, at least one additional unknown autosomal factor must be present in D. simulans that is absent in D. mauritiana (Barbash 2007). These considerations suggest that some components of the Nup96 hybrid incompatibility also evolved after the split of the D. simulans clade species. It appears, then, that both Nup160 and Nup96 hybrid incompatibilities evolved well after the species split of D. melanogaster and the D. simulans clade ancestor and therefore were inconsequential to any reproductive isolation realized in natural populations.
The Nup160 and Nup96 hybrid incompatibilities evolved at similar times, have comparable hybrid lethal effects among the three D. simulans clade species, are both part of complex multicomponent hybrid incompatibilities, and produce proteins predicted to interact directly at the NPC. It is therefore tempting to speculate that these two hybrid incompatibilities are not independent. While Nup160 and Nup96 hybrid incompatibilities may have evolved for similar, nonindependent reasons—most simply, e.g., as incidental by-products of NPC evolution (see later)—their hybrid lethal effects appear genetically independent in two ways. First, the lethality of our double-mutant hybrid males shows that Nup160sim does not require the presence of Nup96mel, nor does Nup96sim require the presence of Nup160mel (Table 3). Second, in an otherwise purely D. melanogaster genetic background, homozygous (or hemizygous) Nup160sim is not lethal in Nup96sim/Nup96mel heterozygotes, and homozygous (or hemizygous) Nup96sim is not lethal in Nup160sim/Nup160mel heterozygotes (Sawamura et al. 2014). The latter findings would seem to rule out the possibility that Nup96sim is the dominant autosomal factor required for Nup160-mediated hybrid lethality (and vice versa). It is possible that a different autosome-encoded NPC protein, perhaps one of the other NUP107 subcomplex proteins or its interactors, is required for Nup160- and Nup96-mediated hybrid lethality (see also Sawamura et al. 2014).
Previous population genetics analyses showed that Nup160 experienced parallel bouts of recurrent adaptive protein evolution in D. melanogaster and, separately, in D. simulans (Tang and Presgraves 2009) (Table 5). The present analyses further suggest that Nup160 experienced recurrent positive selection in the D. simulans clade ancestor (earlier than 240,000 years ago) and in the D. simulans lineage following the split from D. mauritiana and D. sechellia (later than 240,000 years ago). There is no evidence for recurrent positive selection in D. mauritana, which has evolved very slowly (only a single mappable nonsynonymous substitution), or in D. sechellia, which has an order of magnitude less variability than the other species. Why Nup160, Nup96, and other nucleoporins have evolved rapidly (Begun et al. 2007; Presgraves and Stephan 2007; Langley et al. 2012; Nolte et al. 2013; Garrigan et al. 2014) remains unclear. Nucleoporins interact with retroviruses and retrotransposons (Irwin et al. 2005; Dennis et al. 2012; Le Sage and Mouland 2013; Marini et al. 2015), suggesting the opportunity for antagonistic co-evolution with pathogens and/or selfish genetic elements (Presgraves and Stephan 2007). Furthermore, the NPC, along with other nuclear transport proteins, may have evolved in response to segregation distortion in the male germ line (Presgraves 2007; Tracy et al. 2010; Phadnis et al. 2012). There is, however, reason to doubt earlier suggestions that nucleoporins of the NUP107 subcomplex evolved to suppress or compensate for the meiotic drive of selfish centromeres in the female germ line (Presgraves and Stephan 2007; Sawamura 2012): the NUP107 subcomplex in Drosophila, unlike in mammals, does not localize to centromeres or kinetochores (Katsani et al. 2008). Whatever the cause of recurrent evolution at Nup160, the present data suggest that D. melanogaster and the D. simulans clade ancestor inherited some unresolved genetic conflict from their common ancestor. In the D. simulans lineage but not in the D. mauritiana and D. sechellia lineages, this conflict involved nonsynonymous substitutions at Nup160. The lack of evidence for a hard selective sweep in D. simulans may indicate that the conflict (or at least the role of Nup160 in the conflict) has been quiescent during the recent past or, perhaps more likely, that the sweeps were soft. Given the history of natural introgression between the D. simulans clade species (Garrigan et al. 2012), we can further surmise either that the agent(s) of conflict was not exported from D. simulans into its two sister species via migration or that resolution of the conflict in D. mauritiana and D. sechellia involved other genes.
The biological basis of Nup160-mediated hybrid lethality is still unclear. The hybrid lethality of Nup160 is not due to haploinsufficiency because hybrids homozygous for Nup160sim are inviable (Sawamura et al. 2004, 2010). Furthermore, the hybrid lethality of Nup160 is not due to specific suppression of Lhr rescue because Hmr-rescued males also die (Table 1). This conclusion is strengthened by a difference in lethal phase: the Hmr-Lhr hybrid incompatibility kills late larvae, whereas the Nup160 hybrid incompatibility kills pupae (see also Maehara et al. 2012). Sawamura and colleagues have shown that Nup160 also causes female sterility and, among escapers of hybrid lethality, developmental delay and morphological defects (Sawamura et al. 2010; Maehara et al. 2012). This broad range of phenotypes suggests that fundamental cellular functions are compromised by the Nup160 hybrid incompatibility. It will be of interest to determine whether hybrid lethality results from disruption of an essential nucleoporin-mediated function—e.g., nuclear transport, gene expression, and the regulation of chromatin—or some novel gain-of-function hybrid phenotype.
Acknowledgments
We thank Lori Wright for technical assistance; Victoria Cattani, Pierre Gerard, and Amanda Larracuente for helpful discussions; and David Stern for generously sharing unpublished fly reagents. This work was supported by funds from the National Institutes of Health (R01-GM079543) and the David and Lucile Packard Foundation to D.C.P.
Footnotes
Communicating editor: D. J. Begun
Literature Cited
- Araripe L., Eckstrand N., Hartl D. L., Tao Y., 2006. Flanking regions of P-elements inserted in the 3rd chromosome of Drosophila mauritiana. Drosoph. Inf. Serv. 89: 54. [Google Scholar]
- Bapteste E., Charlebois R. L., MacLeod D., Brochier C., 2005. The two tempos of nuclear pore complex evolution: highly adapting proteins in an ancient frozen structure. Genome Biol. 6: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash D. A., 2007. Nup96-dependent hybrid lethality occurs in a subset of species from the simulans clade of Drosophila. Genetics 176: 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash D. A., 2010. Ninety years of Drosophila melanogaster hybrids. Genetics 186: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash D. A., Ashburner M., 2003. A novel system of fertility rescue in Drosophila hybrids reveals a link between hybrid lethality and female sterility. Genetics 163: 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash D. A., Awadalla P., Tarone A. M., 2004. Functional divergence caused by ancient positive selection of a Drosophila hybrid incompatibility locus. PLoS Biol. 2: e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash D. A., Lorigan J. G., 2007. Lethality in Drosophila melanogaster/Drosophila simulans species hybrids is not associated with substantial transcriptional misregulation. J. Exp. Zool. 308B: 74–84. [DOI] [PubMed] [Google Scholar]
- Begun D. J., Holloway A. K., Stevens K., Hillier L. W., Poh Y. P. et al, 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5: e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N., Rabut G., Bai S. W., van Overbeek M., Beaudouin J., et al. , 2001. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154: 1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau N. J., Flores H. A., Wang J., Maheshwari S., Wang X., et al. , 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295. [DOI] [PubMed] [Google Scholar]
- Capelson M., Liang Y., Schulte R., Mair W., Wagner U., et al. , 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140: 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation. Sinauer, Sunderland, MA. [Google Scholar]
- Davis A. W., Roote J., Morley T., Sawamura K., Herrmann S., et al. , 1996. Rescue of hybrid sterility in crosses between D. melanogaster and D. simulans. Nature 380: 157–159. [DOI] [PubMed] [Google Scholar]
- Dennis S., Sheth U., Feldman J. L., English K. A., Priess J. R., 2012. C. elegans germ cells show temperature and age-dependent expression of Cer1, a Gypsy/Ty3-related retrotransposon. PLoS Pathog. 8: e1002591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1937. Genetics and the Origin of Species. Columbia University Press, New York. [Google Scholar]
- Fay J. C., Wu C.-I., 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree P. M., Barbash D. A., 2009. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7: e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D., Kingan S. B., Geneva A. J., Andolfatto P., Clark A. G., et al. , 2012. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22: 1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D., Kingan S. B., Geneva A. J., Vedanayagam J. P., Presgraves D. C., 2014. Genome diversity and divergence in Drosophila mauritiana: multiple signatures of faster X evolution. Genome Biol. Evol. 6: 2444–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard P. R., Presgraves D. C., 2012. Abundant genetic variability in Drosophila simulans for hybrid female lethality in interspecific crosses to Drosophila melanogaster. Genet. Res. 94: 1–7. [DOI] [PubMed] [Google Scholar]
- Grossman E., Medalia O., Zwerger M., 2012. Functional architecture of the nuclear pore complex. Annu. Rev. Biophys. 41: 557–584. [DOI] [PubMed] [Google Scholar]
- Hadorn E., 1961. Zur Autonomie und Phasenspezifitat der Latalitat von Bastarden zwischen Drosophila melanogaster und Drosophila simulans. Rev. Suisse Zool. 68: 197–207. [Google Scholar]
- Hutter P., Ashburner M., 1987. Genetic rescue of inviable hybrids between Drosophila melanogaster and its sibling species. Nature 327: 331–333. [DOI] [PubMed] [Google Scholar]
- Irwin B., Aye M., Baldi P., Beliakova-Bethell N., Cheng H., et al. , 2005. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 15: 641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N., 2010. Hybrid incompatibility genes: remnants of a genomic battlefield? Trends Genet. 26: 317–325. [DOI] [PubMed] [Google Scholar]
- Kalverda B., Fornerod M., 2010. Characterization of genome-nucleoporin interactions in Drosophila links chromatin insulators to the nuclear pore complex. Cell Cycle 9: 4812–4817. [DOI] [PubMed] [Google Scholar]
- Katsani K. R., Karess R. E., Dostatni N., Doye V., 2008. In vivo dynamics of Drosophila nuclear envelope components. Mol. Biol. Cell 19: 3652–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman R. M., Andolfatto P., Coyne J. A., Depaulis F., Kreitman M., et al. , 2000. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156: 1913–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., Stevens K., Cardeno C., Lee Y. C., Schrider D. R., et al. , 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192: 533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand D., Chenel T., Campagne C., Lachaise D., Cariou M. L., 2011. Inter-island divergence within Drosophila mauritiana, a species of the D. simulans complex: Past history and/or speciation in progress? Mol. Ecol. 20: 2787–2804. [DOI] [PubMed] [Google Scholar]
- Le Sage V., Mouland A. J., 2013. Viral subversion of the nuclear pore complex. Viruses 5: 2019–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Hetzer M. W., 2011. Functional interactions between nucleoporins and chromatin. Curr. Opin. Cell Biol. 23: 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lutzmann M., Kunze R., Buerer A., Aebi U., Hurt E., 2002. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21: 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara K., Murata T., Aoyama N., Matsuno K., Sawamura K., 2012. Genetic dissection of Nucleoporin 160 (Nup160), a gene involved in multiple phenotypes of reproductive isolation in Drosophila. Genes Genet. Syst. 87: 99–106. [DOI] [PubMed] [Google Scholar]
- Maheshwari S., Barbash D. A., 2011. The genetics of hybrid incompatibilities. Annu. Rev. Genet. 45: 331–355. [DOI] [PubMed] [Google Scholar]
- Marini B., Kertesz-Farkas A., Ali H., Lucic B., Lisek K., et al. , 2015. Nuclear architecture dictates HIV-1 integration site selection. Nature 14: 227–231. [DOI] [PubMed] [Google Scholar]
- Masly J. P., Jones C. D., Noor M. A. F., Locke J., Orr H. A., 2006. Gene transposition as a novel cause of hybrid male sterility. Science 313: 1448–1450. [DOI] [PubMed] [Google Scholar]
- Matute D. R., Ayroles J. F., 2014. Hybridization occurs between Drosophila simulans and D. sechellia in the Seychelles archipelago. J. Evol. Biol. 27: 1057–1068. [DOI] [PubMed] [Google Scholar]
- McDonald J. H., Kreitman M., 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1940. Bearing of the Drosophila work on systematics, pp. 185–268 in The New Systematics, edited by Huxley. J. S. Clarendon Press, Oxford, UK. [Google Scholar]
- Muller H. J., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Neumann N., Lundin D., Poole A. M., 2010. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE 5: e13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte V., Pandey R. V., Kofler R., Schlotterer C., 2013. Genome-wide patterns of natural variation reveal strong selective sweeps and ongoing genomic conflict in Drosophila mauritiana. Genome Res. 23: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 1996. The unexpected recovery of hybrids in a Drosophila species cross: A genetic analysis. Genet. Res. 67: 11–18. [DOI] [PubMed] [Google Scholar]
- Phadnis N., Hsieh E., Malik H. S., 2012. Birth, death, and replacement of karyopherins in Drosophila. Mol. Biol. Evol. 29: 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., 2003. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 163: 955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., 2007. Does genetic conflict drive molecular evolution of nuclear transport genes in Drosophila? BioEssays 29: 386–391. [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2010a Darwin and the origin of interspecific genetic incompatibilities. Am. Nat. 176(Suppl. 1): 45–60. [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2010b The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11: 175–180. [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., Balagopalan L., Abmayr S. M., Orr H. A., 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423: 715–719. [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., Stephan W., 2007. Pervasive adaptive evolution among interactors of the Drosophila hybrid inviability gene, Nup96. Mol. Biol. Evol. 24: 306–314. [DOI] [PubMed] [Google Scholar]
- Provine W. B., 1991. Alfred Henry Sturtevant and crosses between Drosophila melanogaster and Drosophila simulans. Genetics 129: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyaki P. R., Cuykendall T. N., Wei K. H., Brideau N. J., Kwak H., et al. , 2014. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet. 10: e1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K., 2000. Genetics of hybrid inviability and sterility in Drosophila: the Drosophila melanogaster–Drosophila simulans case. Plant Species Biol. 15: 237–247. [Google Scholar]
- Sawamura K., 2012. Chromatin evolution and molecular drive in speciation. Int. J. Evol. Biol. 2012: 301894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K., Davis A. W., Wu C.-I., 2000. Genetic analysis of speciation by means of introgression into Drosophila melanogaster. Proc. Nat. Acad. Sci. USA 97: 2652–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K., Fujita A., Yokoyama R., Taira T., Inoue Y. H., et al. , 1995. Characterization of a reproductive isolation gene, zygotic hybrid rescue, of Drosophila melanogaster by using minichromosomes. Jpn. J. Genet. 70: 223–323. [DOI] [PubMed] [Google Scholar]
- Sawamura K., Karr T. L., Yamamoto M.-T., 2004. Genetics of hybrid inviability and sterility in Drosophila: dissection of introgression of D. simulans genes in D. melanogaster genome. Genetica 120: 253–260. [DOI] [PubMed] [Google Scholar]
- Sawamura K., Maehara K., Keira Y., Ishikawa H. O., Sasamura T., et al. , 2014. A test of double interspecific introgression of nucleoporin genes in Drosophila. G3 4: 2101–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K., Maehara K., Mashino S., Kagesawa T., Kajiwara M., et al. , 2010. Introgression of Drosophila simulans nuclear pore protein 160 in Drosophila melanogaster alone does not cause inviability but does cause female sterility. Genetics 186: 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K., Taira T., Watanabe T. K., 1993. Hybrid lethal systems in the Drosophila melanogaster species complex. I. The maternal hybrid rescue (mhr) gene of Drosophila simulans. Genetics 133: 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1920. Genetic studies on Drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster. Genetics 5: 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Presgraves D. C., 2009. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323: 779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault S., Singer M., Miyazaki W., Milash D. N., Singh B., et al. , 2004. A complementary transposon toolkit for Drosophila melanogaster. Nature Genetics 36: 283–287. [DOI] [PubMed] [Google Scholar]
- Thomae A. W., Schade G. O., Padeken J., Borath M., Vetter I., et al. , 2013. A pair of centromeric proteins mediates reproductive isolation in Drosophila species. Dev. Cell 27: 412–424. [DOI] [PubMed] [Google Scholar]
- Tracy C., Rio J., Motiwale M., Christensen S. M., Betran E., 2010. Convergently recruited nuclear transport retrogenes are male biased in expression and evolving under positive selection in Drosophila. Genetics 184: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True J. R., Mercer J. M., Laurie C. C., 1996. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142: 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T. K., 1979. A gene that rescues the lethal hybrids between Drosophila melanogaster and D. simulans. Jpn. J. Genet. 54: 325–331. [Google Scholar]