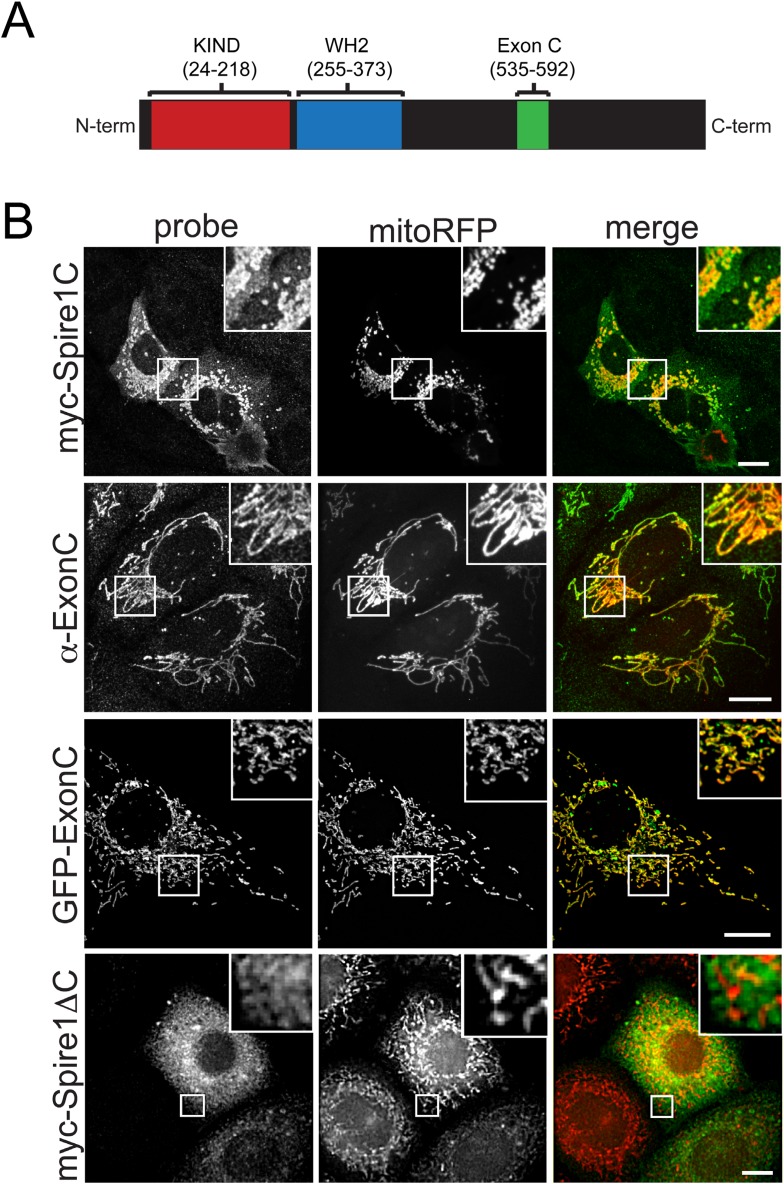
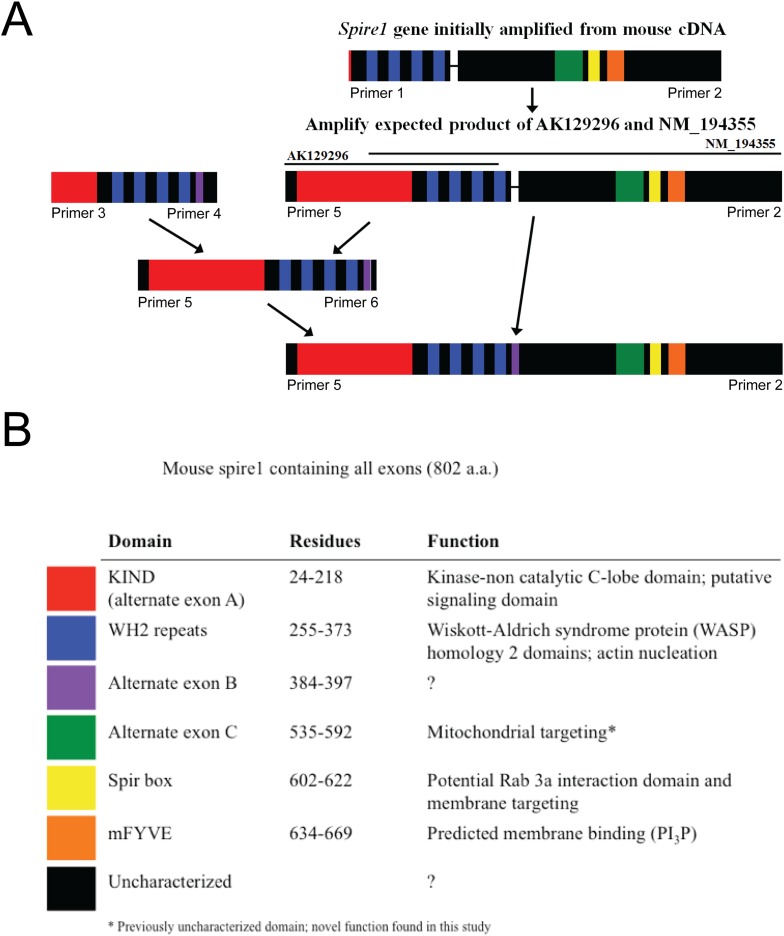
Figure 1. The Spire1 alternate exon ExonC is necessary and sufficient for localization to mitochondria.
(A) Full length Spire1C domain structure: The number ranges indicate the amino acid regions of the conserved domains probed in this study. (B) Spire1C localizes to mitochondria. myc-Spire1C: U2OS cells cotransfected with myc-Spire1C and mitoRFP show robust localization of myc-Spire1C to mitochondria. α-ExonC: U2OS cells stained with an antibody raised against ExonC (α-ExonC) and expressing mitoRFP show endogenous Spire1C labeling on mitochondria. GFP-ExonC: U2OS cells cotransfected with GFP-ExonC and mitoRFP show robust targeting of GFP-ExonC to mitochondria. myc-Spire1ΔC: U2OS cells cotransfected with myc-Spire1ΔC and mitoRFP show no specific targeting of myc-Spire1ΔC to mitochondria. All cells were fixed and primary antibodies were counterstained with Alexa-488 secondary antibody before imaging with confocal fluorescence microscopy. Scale bars: 10 μm. Inserts are magnifications of the boxed regions.