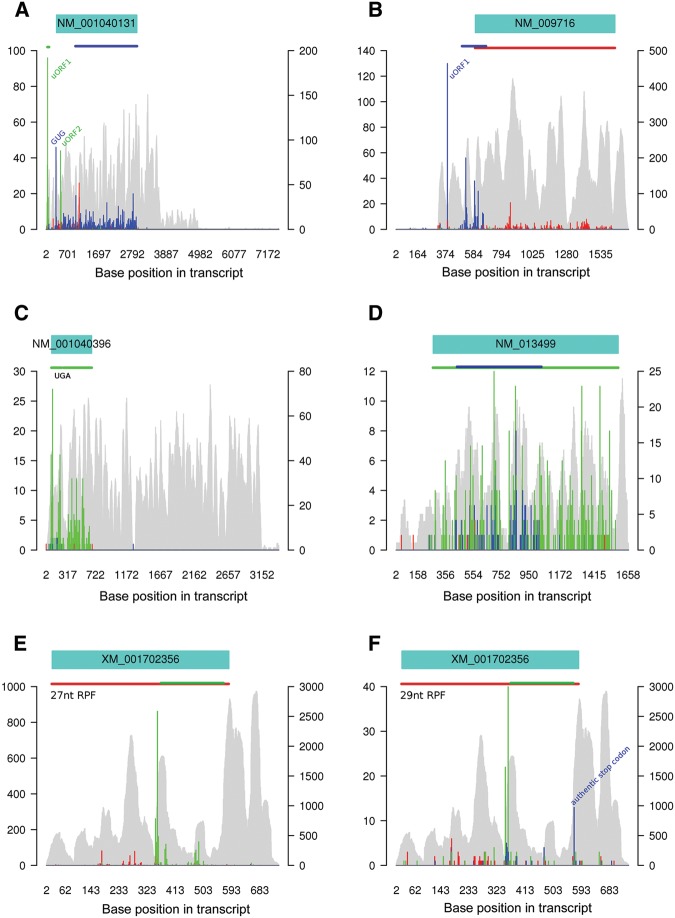
FIGURE 6.
Translated uORFs and overlapping ORFs viewed using riboSeqR. (A) Analysis of RPFs mapping to NCBI RefSeq mRNA NM_001040131 (mouse eIF4G2), showing examples of uORFs. Histograms of the 5′ ends of RPFs (colored) and RNA-seq (gray) reads are shown. The three reading frames to which the 5′ ends of RPFs may map to (relative to nt 1 of the reference sequence) are indicated by different colors, as in Figure 4B. The positions of putative ORFs with at least 50 RPFs mapping to at least 10 locations are shown immediately above the histograms, color-coded as appropriate. A 15-codon sliding window mean of filtered (see Materials and Methods) Ribo-seq CHX RPF counts is shown below the transcript map. The main coding ORF for this transcript is in frame 2 (blue), but translation initiates upstream of the annotated ORF (at the indicated GUG codon). Two uORFs are apparent in frame 1 (green) and an overlapping ORF is also observed (red, see text). (B) Analysis of NM_009716 (mouse ATF4) reveals the two regulatory uORFs (see text), uORF1 and uORF2 (blue), upstream of the main coding sequence (red). (C) Analysis of NM_001040396 (mouse Sel T) showing an example of translation beyond a canonical stop codon by selenocysteine insertion at an in-frame UGA codon (indicated). Note this gene has proportionately a very long 3′ UTR. (D) An example of a likely internal overlapping gene from NM_013499 (Cr1l). (E,F) riboSeqR analysis of Rubisco expression as an example demonstrating a sequencing error in the RefSeq. This error results in an apparent change of reading frame of the 27-nt RPF ribosome profile. The lack of frame 1 (red) reads within the internal ORF and the clear detection of the de facto termination peak at position 568 (using the 29-nt data; F) confirms the RefSeq error.