FIGURE 1.
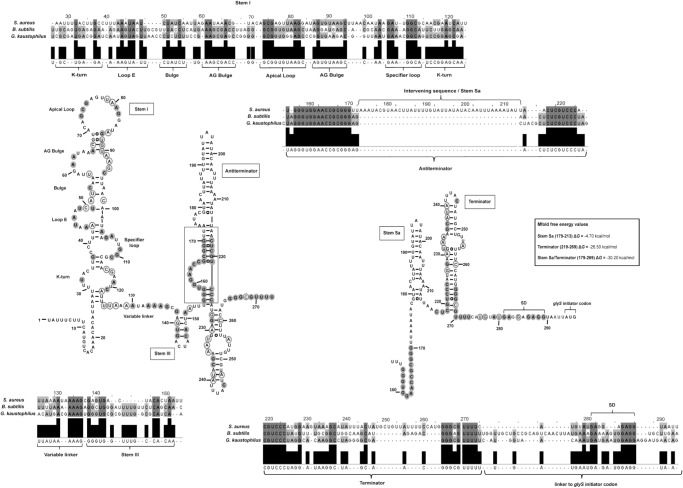
Proposed S. aureus glyS T-box secondary structure based on in silico and biochemical analysis. For the alignment, highly conserved (100%) nucleotides are indicated in dark gray filled circles and moderately conserved (66%) appear in light gray open circles. Alignment numbering corresponds to S. aureus glyS leader sequence. Values corresponding to the thermodynamic stability of the terminator region were calculated according to Mfold free energy prediction tool and appear in a box for each structural element. Annotation numbers: SA1394 S. aureus N315; BSU25270 B. subtilis 168; GK3430 G. kaustophilus HTA426.