Fig 3. Immuno-FISH based karyotype of D-genome chromosomes of wheat and their derived telosomes using CENH3 (white), CRWs (red) and pAs1 (green) as probes.
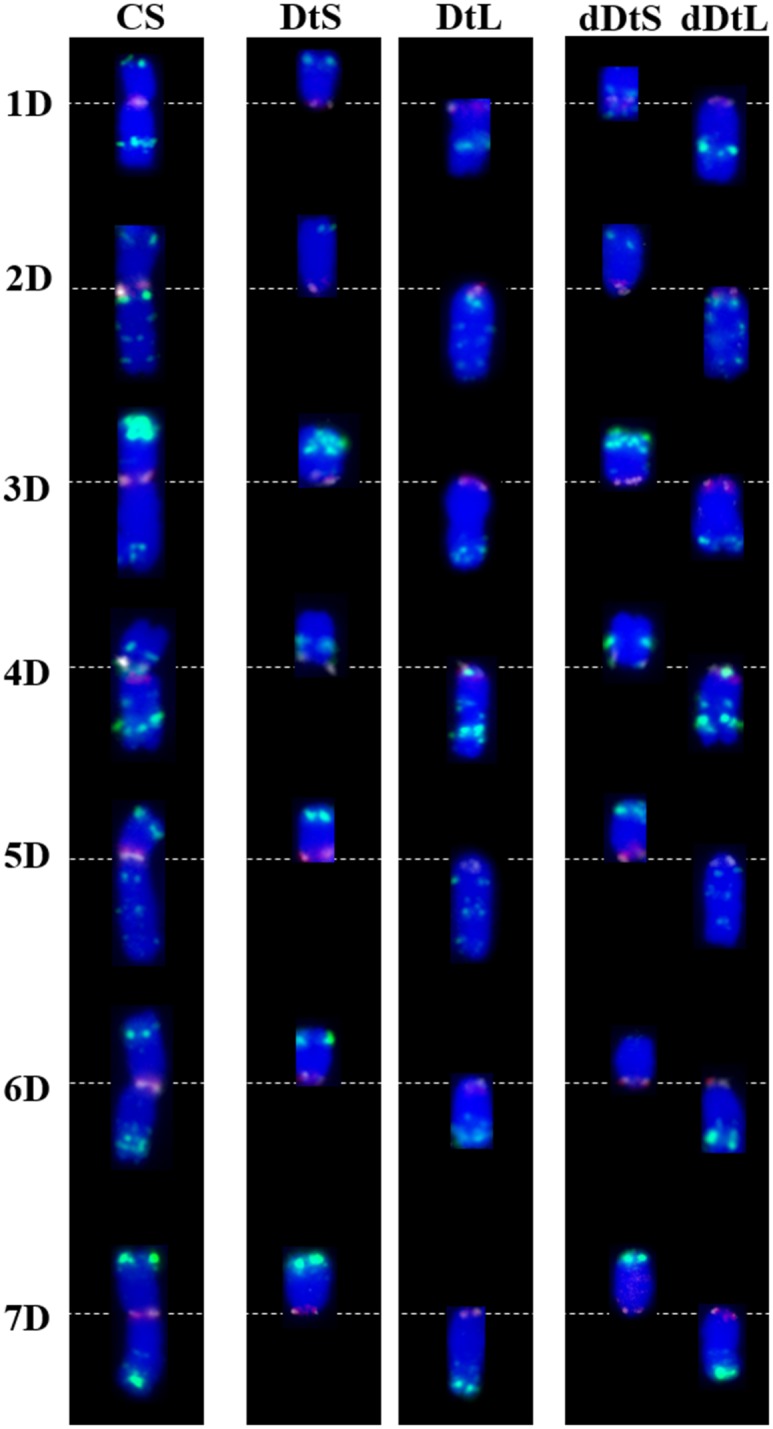
CRWs (red signals) co-localized with CENH3 (white signals) in most of the chromosome except dDt1DS, 4D, Dt4DS, dDt4DS, Dt5DL and dDt5DL. The centromeric regions of chromosome or chromosome arm were seen as pinkish red colors because the CRWs (red signals) are abundant in centromeric region and much brighter than CENH3 signals except in the above mentioned telosomes. The dDt1DS stock contained multiple chromosome rearrangement including inversion, deletion and centromere shift. Note that the CRWs were not detected in Dt4DS and dDt4DS, instead the pAs1 signal was overlapped with the CENH3 signal in these telosomes. A very faint pAs1 FISH site was detected in the terminal region of dDt6DS, indicating a terminal deletion. Short arm and long arm telosomes present in the ditelosomic stocks are represented as (DtS) and (DtL), respectively and short arm and long arm telosomes present in the double ditelosomic stocks are represented as (dDtS) and (dDtL), respectively.
