Fig. 5.
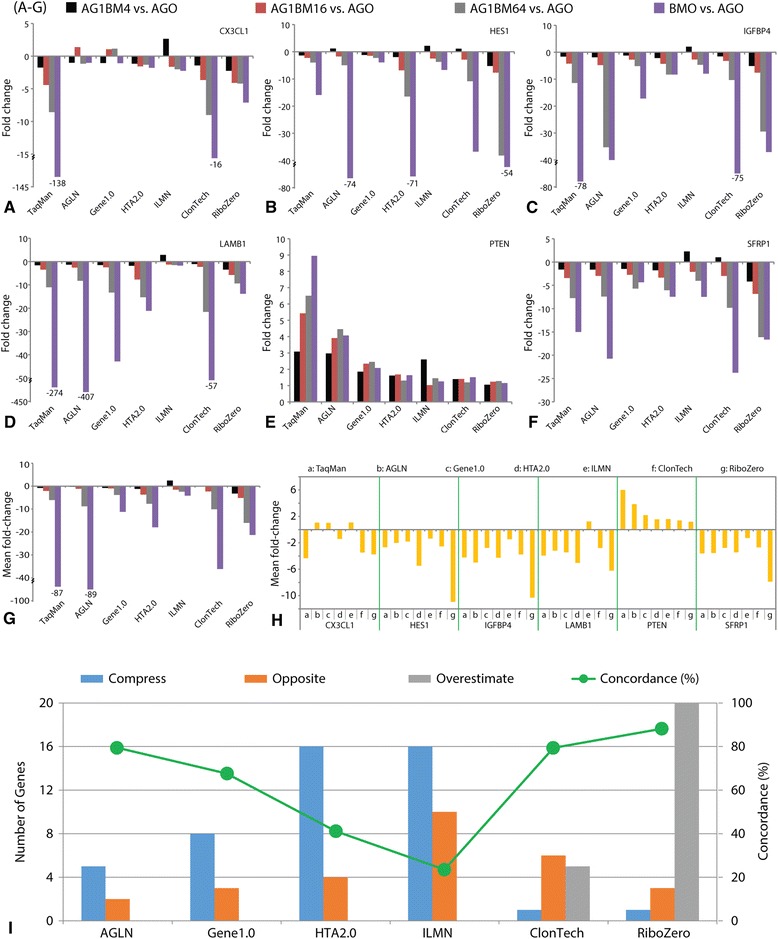
TaqMan qRT-PCR validation. (a-f) fold-change data derived from titrated samples. 6 genes were selected because the TaqMan probes targeted the same transcript pattern as did the microarray probe(set)s; (g) mean fold-change of the 6 genes across platforms; extreme values in (a-d) and (g) were indicated with broken y-axis and actual data; (h) mean fold-change of the 4 titrations across platforms; (I) Concordance of fold-change between TaqMan qPCR (X) and microarrays/RNA-seq protocols (Y), 4 different calls were made: compress, opposite, overestimate, and concordant. When two compared fold-changes are in the same direction but the ratio of X/Y greater than or equal to 2, a value of “compressed” is assigned. Similarly, if the fold-change ratio of X/Y is less than or equal to 0.5 the comparison is deemed “overestimate”. Fold-change ratios between these values are deemed “concordant”. When two fold-changes are not in the same direction and either of them is greater than 2 or less than 0.5, the comparison is determined to be “opposite”. Concordance rates were calculated by number of genes with “concordant” and “overestimate” calls divided by the total genes analyzed
