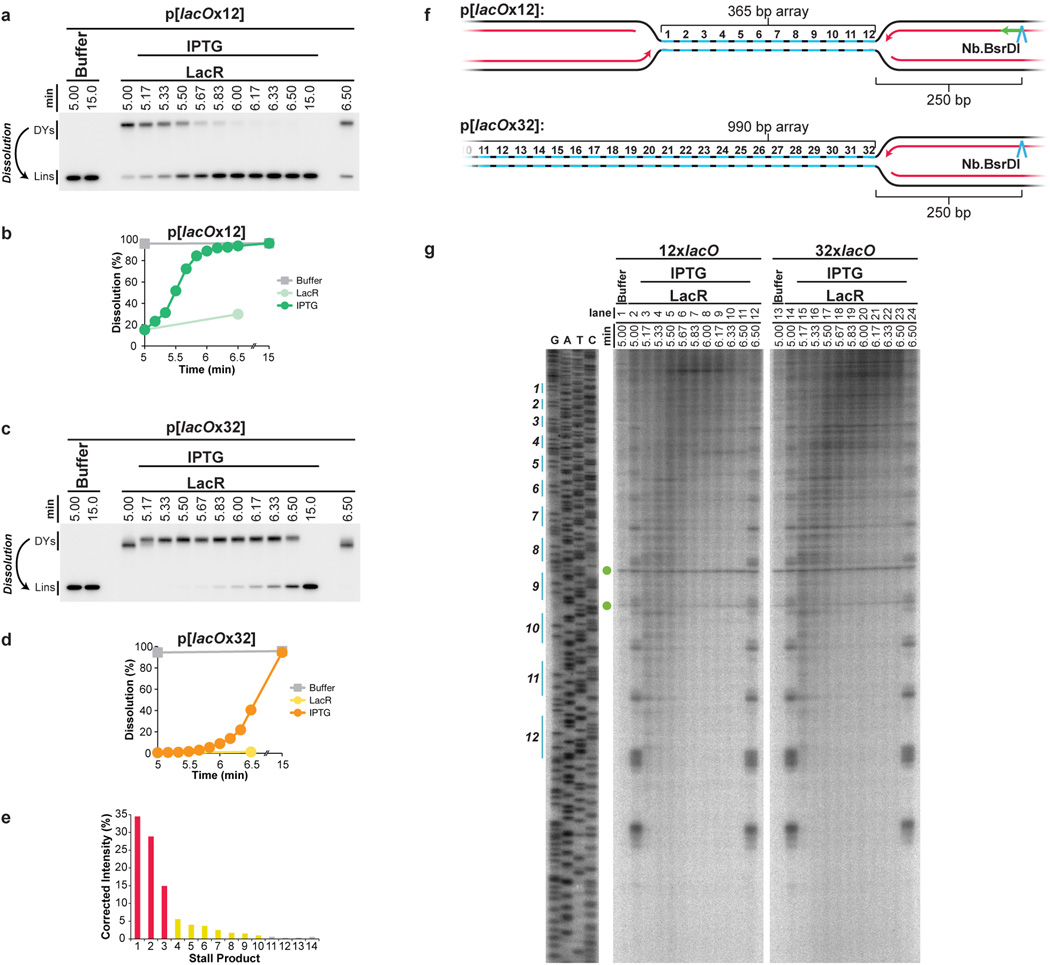
Extended Data Figure 6. Replisome progression through 12x and 32x lacO arrays.
(A–D) To test whether replisomes meet later in a lacOx32 array than a lacOx12 array, we monitored dissolution. LacR Block-IPTG release was performed on p[lacOx12] and p[lacOx32] and radiolabelled termination intermediates were digested with XmnI to monitor the conversion of double-Y molecules to linear molecules (Dissolution). Cleaved molecules were separated on a native agarose gel, detected by autoradiography (A,C), and quantified (B,D). Upon IPTG addition, dissolution was delayed by at least 1 minute within the 32xlacO array compared to the 12xlacO array (B,D). Moreover, by 6 minutes, 92% of forks had undergone dissolution on p[lacOx12] while only 9% had dissolved on p[lacOx32] (B,D).
(E) Stall products within the 12xlacO array (Fig. 3B, Lane 2) were quantified, signal was corrected based on size differences of the products, and the percentage of stall products at each stall point was calculated. 78% of leading strands stalled at the first three arrest points (red columns), 19% stalled at the fourth to tenth arrest points (yellow columns) and the remaining 3% stalled at the tenth to fourteenth arrest points (grey columns). The appearance of fourteen arrest points reproducible and surprising, given that the presence of only twelve lacO sequences was confirmed by sequencing in the very preparation of p[lacOx12] that was used in Fig. 3. The thirteenth and fourteenth arrest points cannot stem from cryptic lacO sites beyond the twelfth lacO site, as this would position the first leftward leading strand stall product ∼90 nucleotides from the lacO array, instead of the observed ∼30 nucleotides (see F-G). At present, we do not understand the origin of these stall products.
(F–G) Progression of leftward leading strands into the array. The same DNA samples used in Fig. 3 were digested with the nicking enzyme Nb.BsrDI, which released leftward leading strands (F), and separated on a denaturing polyacrylamide gel (G). The lacO sites of p[lacOx12] are highlighted in blue on the sequencing ladder (G), which was generated using the primer JDO109 (Green arrow, F). Green circles indicate two non-specific products of digestion. These products arise because nicking enzyme activity varies between experiments, even under the same conditions. There was no significant difference in the pattern of leftward leading strand progression between the 12xlacO and 32xlacO arrays, as seen for the rightward leading strands (Fig. 3B). Specifically, by 5.67 minutes, the majority of leading strands had extended beyond the seventh lacO repeat within lacOx12 (lane 6) and the equivalent region of lacOx32 (lane 18). Therefore, progression of leftward leading strands is unaffected by the presence of an opposing replisome, suggesting that converging replisomes do not stall when they meet.