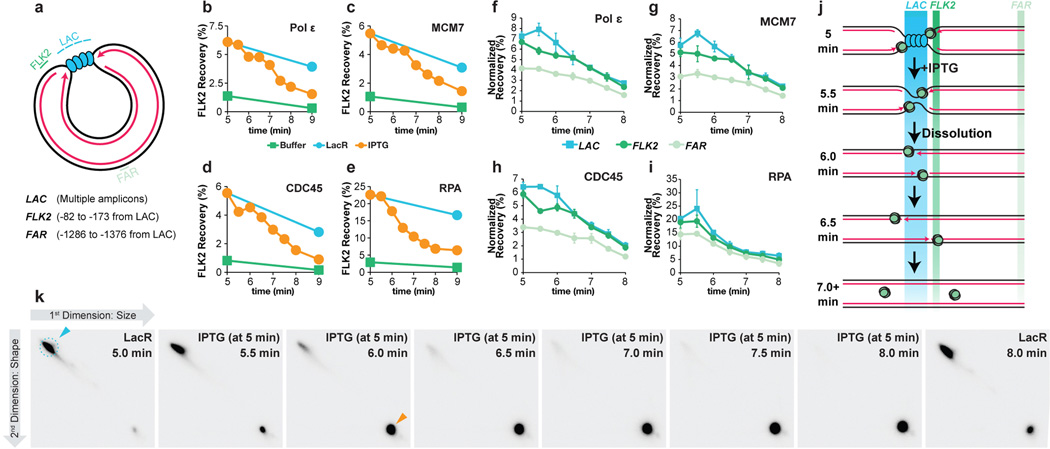
Extended Data Figure 7. Supplemental ChIP data.
(A) Cartoon depicting the LAC, FLK2 and FAR loci, which were used for ChIP. Their precise locations relative to the leftward edge of the lacO array are indicated. The LAC amplicon is present in four copies distributed across the lacOx16 array and three copies distributed across the lacOx12 array.
(B–E) p[lacOx12] was incubated with buffer or LacR and termination was induced at 5 minutes by IPTG addition. MCM7, RPA, CDC45, and Polε ChIP was performed at different time points after IPTG addition but also in the buffer control and no IPTG control. Recovery of FLK2 was measured as a percentage of input DNA. Upon IPTG addition, ChIP signal declined and by 9 minutes was comparable to the buffer control, demonstrating that unloading of replisomes was induced within 4 minutes of IPTG addition.
(F) To test whether movement of the replisome into and out of the lacO array could be detected upon IPTG addition, termination was monitored within a lacO array, and we performed ChIP of the leading strand polymerase Polε, which was inferred to move into and out of the array based on the behavior of leading strands during termination (Extended Data Fig 2B–E). It was predicted that Polε ChIP at the LAC locus should increase slightly as Polε enters the lacO array and decline again as converging polymerases pass each other, but persist at FLK2 while the polymerases move out of the array. Prior to IPTG addition, Polε was enriched at LAC and FLK2 compared to FAR, consistent with the leading strands being positioned on either side of the lacO array (Extended Data Fig 2C, Fig. 3). Upon IPTG addition, Polε became modestly enriched at LAC compared to FLK2 (5.5 min) but then declined to similar levels at both LAC and FLK2 by 6.5 min. These data are consistent with the leading strand polymerases entering the lacO array and passing each other.
(G–H) To test whether CMG exhibited the same ChIP profile as Polε, MCM7 and CDC45 ChIP was performed using the same samples. Following IPTG addition, MCM7 and CDC45 were enriched at LAC compared to FLK2 (5.5 min), then declined to similar levels at both LAC and FLK2 by 6.5 min, as seen for Polε (F). These data are consistent with a model in which CMGs enter the array and pass each other during termination. A caveat of these experiments is the relatively high recovery of the FAR locus in MCM7, CDC45, and Polε ChIP. Specifically, signal was at most only ∼2-fold enriched at LAC compared to FAR. This was not due to high background binding, because by the end of the experiment (10 minute time point, not shown), we observed a decrease in signal of ∼5–7 fold. Furthermore, we observed ∼5–7 fold enrichment in binding (ChIP) of replisome components to p[lacOx12] that had been incubated in LacR compared to a buffer control (see G-I, below). Instead, the high FAR signal was likely due to poor spatial resolution of the ChIP. Consistent with this, when a plasmid containing a DNA interstrand cross-link (ICL) was replicated, essentially all replisomes converged upon the ICL but the ChIP signal for MCM7 and CDC45 was only ∼3–4 fold enriched at the ICL compared to a control locus41. We speculate that the higher background observed at the control locus in our experiments is due to the decreased distance of the control locus from the experimental locus (1.3 kb for p[lacOx16] and p[lacOx12] vs. 2.4 kb for the ICL plasmid) and possibly due to increased catenation of the parental strands during termination. The high signal at FAR should not complicate interpretation of the MCM7, CDC45 and Polε ChIP (F), as signal at FAR was essentially unaltered between 5 and 6.5 minutes. Further evidence that the high signal seen at the FAR locus emanates from forks stalled near the lacO array is presented in panel (K).
(I) ChIP of RPA was performed on the same chromatin samples used in B-D. As seen for pol ɛ, MCM7, and CDC45, enrichment of RPA at LAC compared to FAR was relatively low, consistent with poor spatial resolution.
(J) Predicted binding of CMGs to the LAC, FLK2 and FAR loci before and after IPTG addition if CMGs converging CMG pass each other.
(K) To determine whether most forks stalled at the array and not elsewhere in the plasmid, we performed a time course in which p[lacOx16] undergoing termination was examined by 2-dimensional gel electrophoresis (2-D gel) at various time points. p[lacOx16] was pre-bound to LacR and replicated in Xenopus egg extract containing [α-32P]dATP. Termination was induced by IPTG addition and samples were withdrawn at different times. Radiolabelled replication intermediates were cleaved with XmnI (as in Extended Data Fig. 1A) and separated according to size and shape on 2-D gels50. A parallel reaction was performed in which samples were analyzed by ChIP, which was one of the repeats analyzed in (B)-(E). In the presence of LacR, a subset of Double Y molecules accumulated (blue arrowhead), demonstrating that 83% of replication intermediates (signal in dashed blue circle) contained two forks converged at a specific locus. Following IPTG addition, linear molecules rapidly accumulated (orange arrow) as dissolution occurred. Importantly, the vast majority of signal was present in the discrete double-Y and linear species (blue and orange arrows), demonstrating that the relatively high ChIP signal observed at FAR in panels F-I was derived from forks present at the lacOx16 array and not elsewhere.