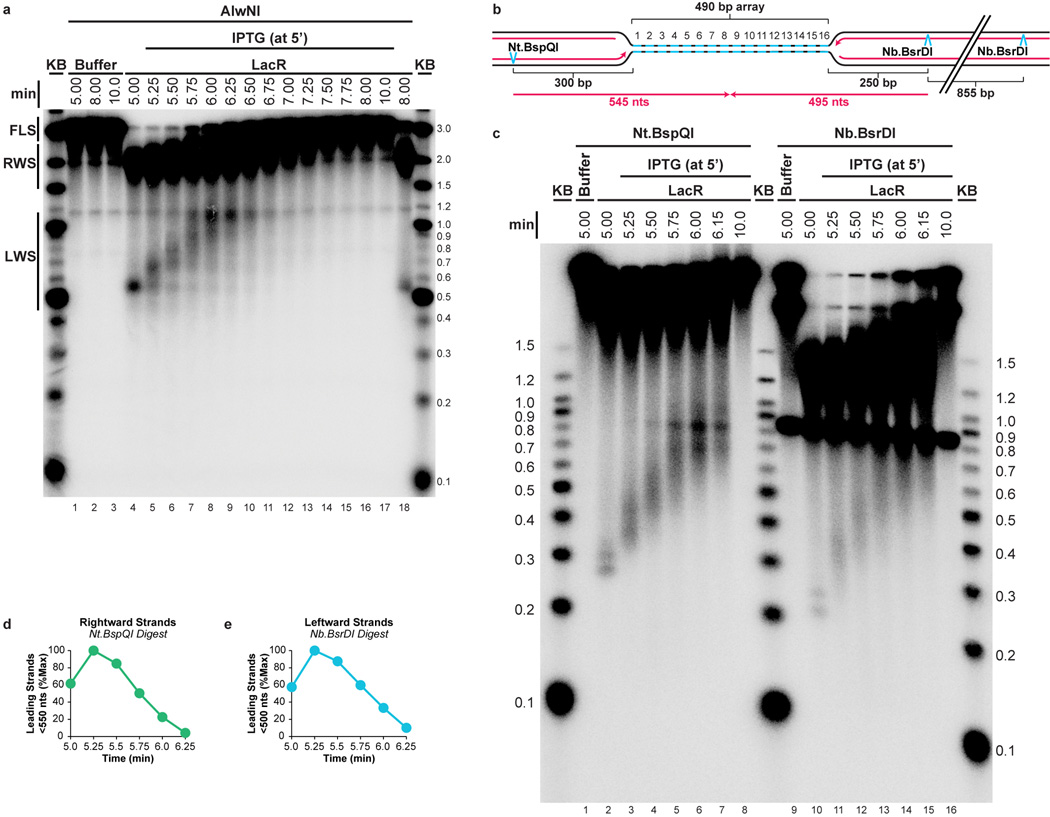
Extended Data Figure 2. Supplemental fork progression data.
(A) The gel shown in Figure 1E was overexposed and shown in its entirety so that the smaller leftward strands (LWS, Fig 1D) could be detected. As observed for the rightward strands (RWS, Fig 1E), LWS rapidly increased in size and then disappeared as they were ligated to produce full length strands (FLS, Fig 1E).
(B–E) To determine whether the heterogeneity of LWS (A) and RWS (Fig 1E) was due to delayed extension of lagging strands, or because a significant fraction of leading strands did not restart upon IPTG addition, we specifically monitored leading strand progression upon IPTG addition on p[lacOx16]. To this end, DNA samples were treated with Nt.BspQI or Nb.BsrDI to specifically liberate the rightward or leftward leading strands, respectively (B), and DNAs were separated on a denaturing agarose gel (C). Prior to IPTG addition, discrete leading strand products of the expected size were observed (lanes 2 and 10). The presence of two stall products reflects the fact that at a slow rate, the replisome bypasses LacR (see also Fig 3). Upon IPTG addition, these species rapidly and completely shifted up the gel, indicating that rightward and leftward leading strands restarted efficiently. Therefore, the heterogeneity of the LWS (A) and RWS (Fig. 1E) is probably due to delayed ligation of a new Okazaki fragment to the lagging strands. Quantification of leading strands that had not reached the midpoint of the array (rightward and leftward strands smaller than 550 and 500 nts, respectively, B) revealed that by 6.25 minutes, 90% of rightward and leftward leading strands passed the midpoint of the array (D–E). This demonstrates that leading strands pass each other when forks meet.