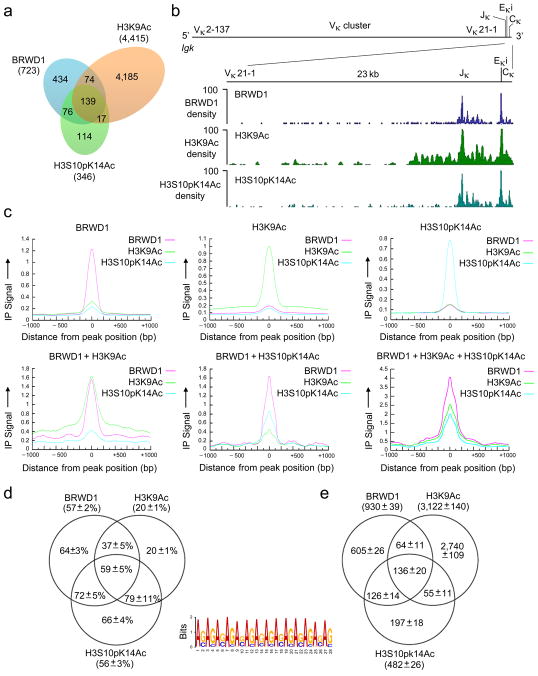
Figure 6.
Recruitment of BRWD1 to H3K9Ac and H3S10pK14Ac genome-wide. (a) Overlap of peaks (P<10−7) obtained by ChIP-Seq for BRWD1, H3K9Ac and H3S10pK14Ac from purified WT small pre-B cells. Data are representative of two independent experiments. (b) ChIP-Seq analysis of the binding of BRWD1, H3K9Ac and H3S10pK14Ac at the Igk locus in purified WT small pre-B cells presented as smoothed density (where ‘density’ indicates sequence ‘read’). Data are representative of two independent experiments. Igk locus shows the location of Vκ, Jκ, Eκi and Cκ gene segments (mm9 chromosome 6: 70,653,572–70,676,748). (c) Alignment of BRWD1, H3K9Ac and/or H3S10pK14Ac enrichment in ChIP-Seq peaks. Y-axis represents normalized immunoprecipitation signal distribution for ChIP peaks centered at 0 bp. (d) Percentage of peaks containing at least one extended GAGA motif (GA11). The +/− errors are standard deviations from 100 bootstrapping runs for each peak group. (e) Total number of extended GAGA motif (GA11) hits in different ChIP-Seq groups. The +/− errors are standard deviations from 100 bootstrapping runs for each peak group.