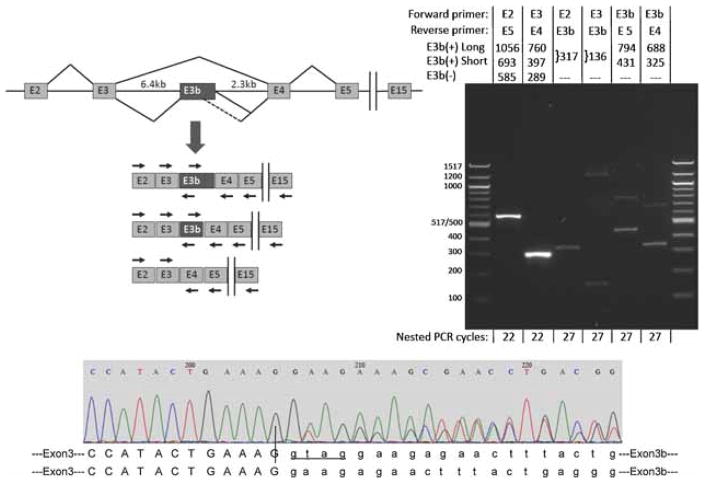
Figure 3.
Alternative splicing of SLC6A3 exon E3b in post-mortem human substantia nigra. Top left panel: Schematic (not drawn to scale) of the region comprising exons 2 through 15 of the primary SLC6A3 transcript and spliced mRNAs. Vertical bars indicate the omission of exons 6–14 from the diagram. The diagram illustrates skipping and inclusion of E3b and use of two alternative E3b 5′ splice sites spaced 363bp apart. The locations and orientations (forward or reverse) of primers used for RT-PCR are shown by arrows above or below the corresponding exons (primer sequences are given in Supplementary Methods Section). Note that the intron regions flanking E3b measure 6.4 and 2.3 kb, respectively, precluding amplification of unprocessed pre-mRNA with any of the primer combinations under the PCR conditions used. Top right panel: RT-PCR analysis of E3b splicing. mRNA from normal adult substantia nigra substantia nigra was reverse-transcribed with oligo-dT primer and the cDNA was subjected to 20 cycles of PCR with primers against exon E2 (E2.F) and E15 (E15.R). The products were diluted and subjected to re-amplification with different combinations of nested primers targeting exons E2 through E5. The primer combinations for the nested PCR and the expected sizes of amplimers corresponding to E3b(+) and E3b(−) isoforms are indicated above the gel lanes; E3b(+) long and E3b(+) short correspond to use of the distal or proximal 5′ splice sites for E3b, respectively. The sizes of E3b(+) amplimers shown correspond to use of the first of the two tandem 3′ splice sites; use of the second 3′ splice site shortens the products by four nt (see bottom panel). The number of cycles used in the nested PCR reaction is indicated below each lane. Note that in lanes 2 (E2 + E5 primers) and 3 (E3 + E4 primers), the E3b(+) amplimers are not visible because they are much less abundant than the E3b(−) amplimers. The identities of amplimers incorporating exon E3b were verified by sequencing. The identities of the large amplimers in lanes 3 (faint) and 4 have not been investigated. The same results were obtained when the reverse primer in the primary amplification targeted exon E5, E13, or E15. When the primary PCR was performed on the same RNA samples without reverse transcription, no amplimers were observed with any of the nested PCR reactions using the same number of cycles (not shown). Bottom panel: Sequencing E3b(+) amplimers across the E3/E3b junction shows that both of the tandem 3′ splice sites for exon E3b are used. E3b begins at nucleotide 206 in this representative sequencing trace. Upstream of this position the sequence trace is homogeneous (corresponding to the end of exon E3), but downstream it is a mixture of two sequences that correspond to use of the two different 3′ splice sites to begin E3b. The two predicted E3/E3b junction sequences are aligned below the trace. Nucleotides in lower case mark the beginning of the alternatively spliced cassette exon E3b. Four additional nucleotides (GTAG) are spliced out as part of the intron when the downstream site is used (lower sequence).