Figure 3.
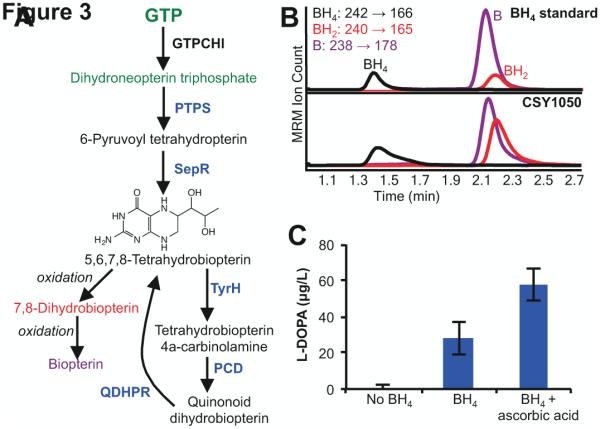
Production of the TyrH cosubstrate BH4 in engineered yeast. (A) Engineered biosynthetic pathway for production and recycling of BH4. Enzymes labeled in black indicate endogenous yeast enzymes, blue indicate heterologous enzymes. Compounds labeled in green represent the connection between yeast central metabolism and BH4 biosynthesis. GTPCHI, GTP cyclohydrolase I; PTPS, 6-pyruvoyl-tetrahydropterin synthase; SepR, sepiapterin reductase; PCD, pterin-4alpha-carbinolamine dehydratase (PCD); QDHPR, quinonoid dihydropteridine reductase. (B) BH4 production in yeast. Top, BH4 standard analyzed with LC-MS/MS in MRM mode. Bottom, CSY1050 (CSY1039 with BH4 biosynthesis genes integrated) media analyzed for BH4, BH2, and B with LC-MS/MS in MRM mode. Traces are color coded to match compound names in Fig. 3A. (C) L-DOPA production as a function of BH4 and ascorbic acid. CSY1002 (no BH4) and CSY1050 (BH4 and BH4 + ascorbic acid) each express the wild-type RnTyrH (pCS3231). Strains were grown in selective media lacking tyrosine for 96 hours before analysis. CSY1050 was grown in the presence and absence of 2 mM ascorbic acid. Data is reported as the mean ± s.d. of at least 3 independent experiments.
