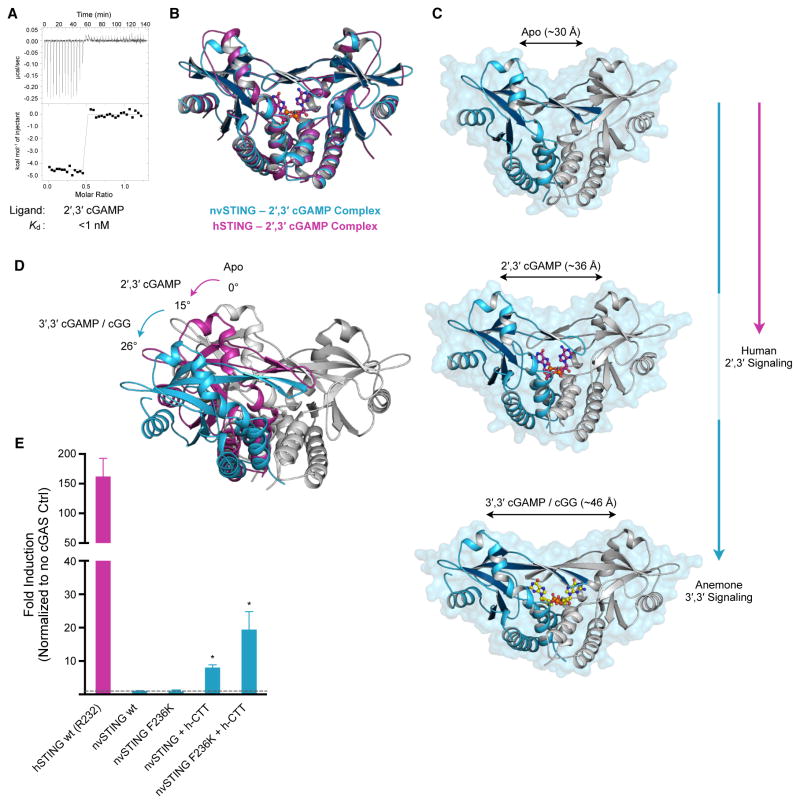
Figure 5. 2′,3′ cGAMP Traps a Unique and Conserved STING Conformation.
(A) ITC measurements of nvSTING affinity for 2′,3′ cGAMP. (B) Structural overlay of nvSTING–2′,3′ cGAMP complex (blue) and hSTING–2′,3′ cGAMP complex (magenta) (PDB 4KSY) demonstrating the monomer wing rotation and core CDN-interacting portion of nvSTING and hSTING are unchanged. (C) Structural comparison of nvSTING structures in various ligandbound complexes. CDN ligands lock STING in alternative conformations as measured by the distance between the apical monomer wing domains (monomer 1 in blue, monomer 2 in grey). nvSTING–3′,3′ CDN interactions result in complete monomer rotation (blue vertical line) while primary hSTING–2′,3′ cGAMP signaling traps a partially rotated intermediate structure (magenta line). (D) Endogenous anemone 3′,3′ second messengers trigger an ~26° rotation in monomer wing domains from the apo state (apo grey, 3′,3′-bound in blue), while human 2′,3′ cGAMP traps an ~15° rotated structural intermediate in the nvST ING and hSTING structures (2′,3′-bound in magenta). (E) Cell interferon β luciferase as in Figure 2A, using indicated STING plasmids stimulated with human cGAS overexpression. Error bars represent the SE of the mean of at least three independent experiments (asterisk denotes p <0.002). ITC data is representative of at least three independent experiments. See also Figure S5 and Table S1.