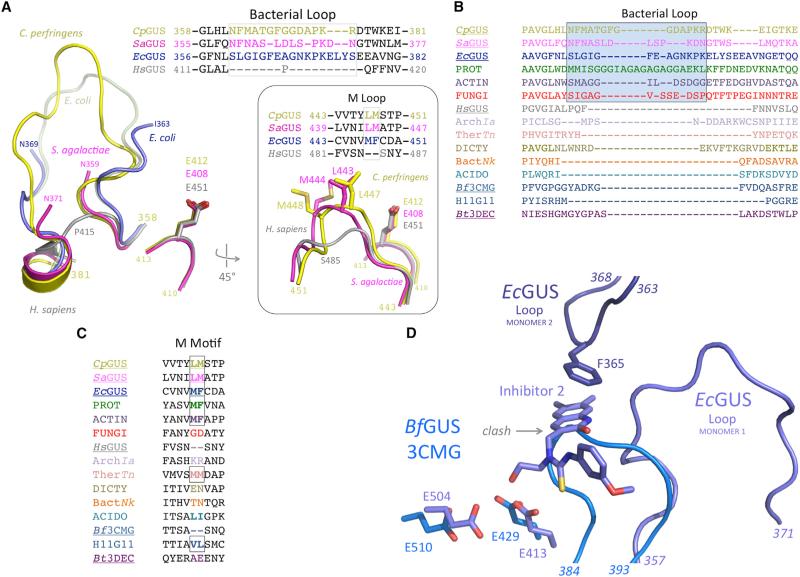
Figure 2. Bacterial- and Mammalian-Type β-Glucuronidases Exhibit Distinct Bacterial and M Loops.
(A) The three novel bacterial β-glucuronidase structures presented here (yellow, magenta, blue) superimposed on the structure of human β-glucuronidase (gray) reveals both the lack of a bacterial loop in the mammalian enzyme, as well as the lack of an M loop in the mammalian enzyme (inset).
(B) The bacterial loop, boxed, is maintained in the L-GUS enzymes, including those from several fungi (red), but is missing from the no-loop β-glucuronidases (NL-GUS) and the B. thetaiotaomicron (Bt3DEC) β-galactosidase. The enzymes for which a crystal structure is available are underlined. PROT, Proteobacteria Vibrio harveyi, WP_005434141; ACTIN, Actinobacteria Corynebacterium massiliense, WP_022863751; FUNGI, Eukaryota Aspergillus niger, CAK36851; ArchIa, Archaea Ignisphaera aggregans, WP_013302863; TherTn, Thermotogae Thermotoga naphthophila, ADA67771; DICTY, Dictyoglomi Dictyoglomus turgidum, ACK42813; BactNk, Bacteroidetes Niastella koreensis, AEV98753; ACIDO, Acidobacteriua Terriglobus roseus, WP_01478374; H11G11, 745-residue protein from uncultured bacterium.
(C) A hydrophobic M loop is missing from several no-loop β-glucuronidases (NL-GUS), such as HsGUS and Bf3CMG, but is present in others, and is conserved in the bacterial loop-containing β-glucuronidases (L-GUS). The enzymes for which a crystal structure is available are underlined.
(D) Active-site superposition of EcGUS-Inh2 complex (dark blue) on BfGUS (3CMG; light blue) showing the catalytic residues and bacterial loop of EcGUS. The 384–393 region of BfGUS, which is significantly shorter than the bacterial loop of EcGUS (357–371), would clash with the binding site of compounds such as Inhibitor 2. In addition, in the EcGUS tetramer, loops from different monomers (e.g., 1 and 2 shown) participate in binding contacts with Inhibitor 2. BfGUS does not form an analogous tetramer and cannot form a similar inhibitor binding site.