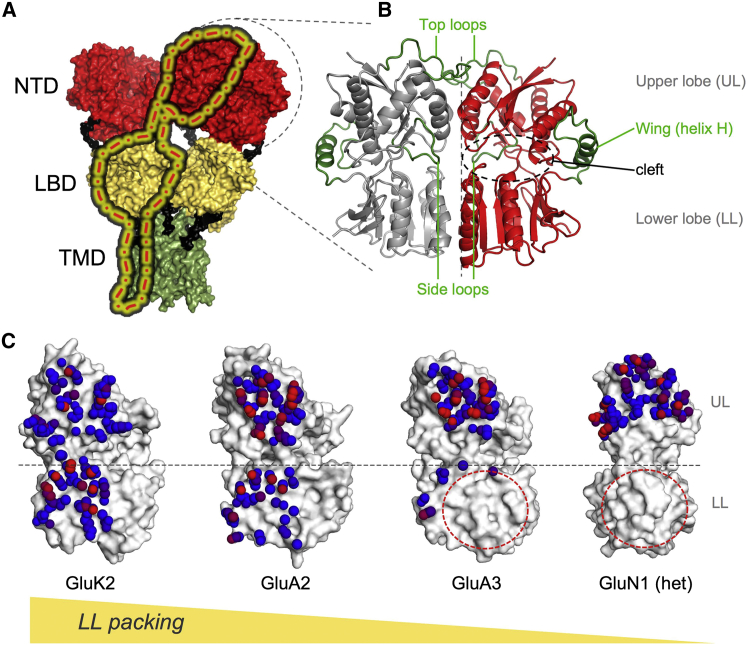
Figure 1.
Structure of iGluR NTDs. (A) A structure of a homomeric GluA2 AMPAR (PDB ID: 3KG2) (18) is shown with the three main domain layers colored red (NTD), yellow (LBD), and green (TMD). The interdomain linkers are colored black. One subunit is highlighted. (B) A zoom-in of an NTD dimer illustrates the clamshell structure of each protomer (upper lobe (UL) and lower lobe (LL) separated by a cleft), as well as four features that vary between iGluRs: the overall dimeric packing, the top loop, the wing element, and the side loop. (C) The dimer interfaces of selected iGluR NTD dimers are shown as color-coded spheres for atoms forming contacts (interfacial atom-to-atom distance = <4.5 Å). Spheres are colored according to the number of contacts, from blue (n = 1) to red (n ≥ 7). A spectrum of LL packing is shown, comparing iGluR NTD dimers (left to right). The kainate receptors (exemplified by a GluK2 homodimer; PDB ID: 3H6H) (60) show the most LL packing similar to their UL packing (these interfaces show local contact densities (LDs) (100) of 27.0 and 35.1, respectively). Among the AMPAR paralogs, homodimeric LL packing correlates with affinity with GluA2 (PDB ID: 3H5V, dimer AB) (46) having the most tightly packed LLs (LD 23.4 vs. 43.5 for UL) and GluA3 (PDB ID: 3O21, dimer CD) (57) having minimal LL packing (LD 6.7 vs. 45.0 for UL). The NMDARs are at the far end of the spectrum, with no LL packing at all (shown for GluN1 of the heterodimer; PDB ID: 3QEL, dimer AB) (64). The dashed red circles in GluA3 and GluN1 highlight the lack of LL packing. A more detailed analysis of NTD dimer interfaces can be found in our recent studies (32, 56, 57). To see this figure in color, go online.