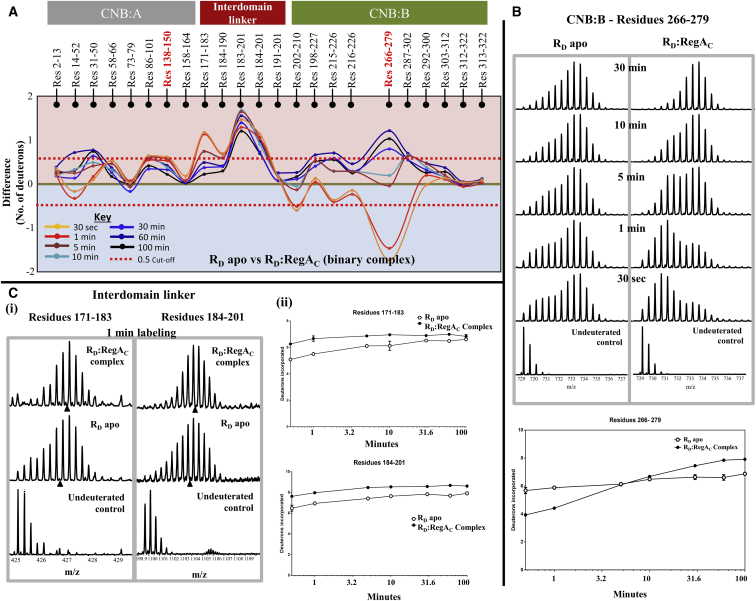
Figure 5.
Mapping transient RD:RegAC interactions by HDXMS. (A) Difference plot, plotting absolute difference in deuterons (y axis) between apo RD and RD:RegAC binary complex for each pepsin fragment peptide listed from the N- to C-terminus (x axis). Points in the negative scale (shaded blue) represent a decrease in deuterium exchange upon RegAC binding, while points in the positive scale (shaded red) represent increases in deuterium exchange upon RegAC binding. Peptides spanning the cAMP binding pocket in CNB:A and B are highlighted (red). Each deuterium labeling time for every peptide is depicted and colored according to key. A difference of ±0.5 Da is considered significant and is shown (red dashed line). Plots were generated using the software DYNAMX. (B) (Top panel) Stacked mass spectra of peptides spanning residues 266–279 in CNB-B domain of RD. Bimodal spectra from apo RD are compared with bimodal spectra from the binary complex. Mass spectra are stacked in order of increasing deuterium-labeling time as shown. (Bottom panel) Deuterium exchange plot comparing apo RD and RD:RegAC for the same peptide is depicted. (C) Representative mass spectra and deuterium exchange plots for two peptides spanning residues 171–183 and 184–201 are shown comparing RD apo state and RD:RegAC state. Semilog deuterium exchange plots are generated with x axis in the log scale and y axis in linear scale; error bars are indicated. All plots are generated in GRAPHPAD PRISM 6.