Figure 3. Conserved and paralogous patches in lincRNAs.
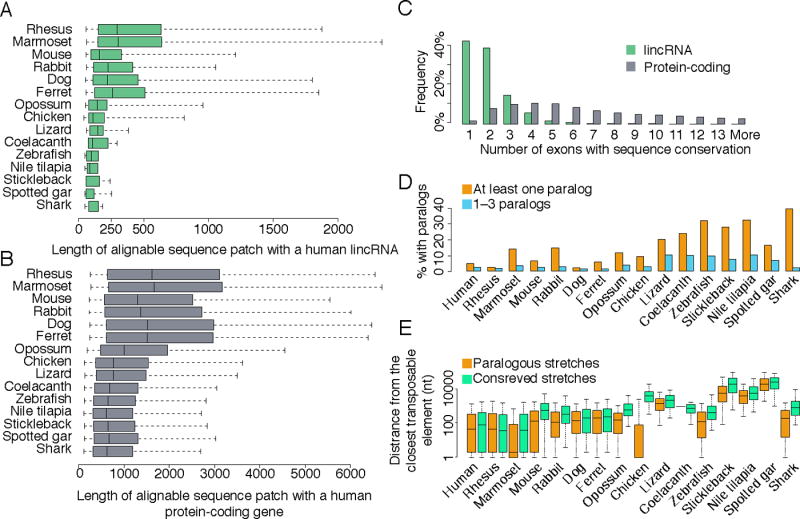
(A) Distributions of lengths of conserved patches, defined as the total length of the sequence alignable by BLASTN between a human lincRNA transcript and any lincRNA transcript in the indicated species. (B) Same as A, but for protein-coding gene reconstructions. (C) When considering patches of conservation of human lincRNAs with species except for rhesus, the distributions of the number of exons that overlap a conserved sequence patch. (D) Fraction of lincRNA genes that have a paralogous lincRNA (BLASTN E-value<10−5) within the same species. Fractions are shown either when including all paralogous pairs, or only considering lincRNAs that have less than four distinct paralogous lincRNAs. (E) Distributions of distances of paralogous and conserved sequence patches from the nearest annotated transposable element. See also Figure S4.
