Figure 1.
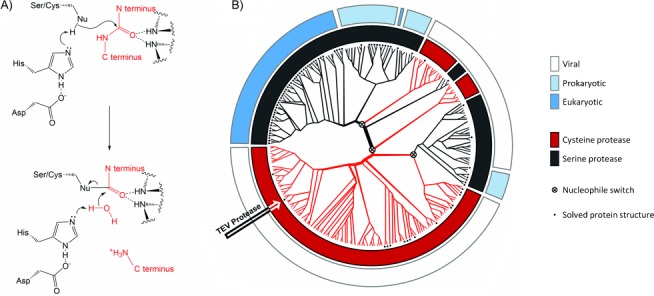
A) Simplified catalytic mechanism of cysteine and serine proteases (Nu=S, e.g., in TEVCys; or Nu=O, e.g., in TEVSer).7 In the enzyme’s catalytic triad (black) aspartate aligns and polarises histidine, which reduces the nucleophile’s pKa and positions it for reaction. The activated nucleophile attacks the carbonyl of the peptide bond of the substrate (red), a tetrahedral intermediate is stabilised by backbone amide hydrogens (“oxyanion hole”), and the C terminus of the substrate is ejected, leaving a covalent adduct behind. This acyl–enzyme intermediate (lower panel) is hydrolysed by histidine-activated water to release the N terminus of the substrate (aided by proton transfer to histidine), and this regenerates the free enzyme. B) The phylogeny of the PA clan (MEROPS classification)11 of proteases with nucleophile and possible nucleophile-switching events indicated. Proteases with known structures were aligned to TEVCys by structural comparison using DALI,13 sequences of unknown structure were added by sequence alignment with BLASTp (see Table S1 and Figure S1 for details).
