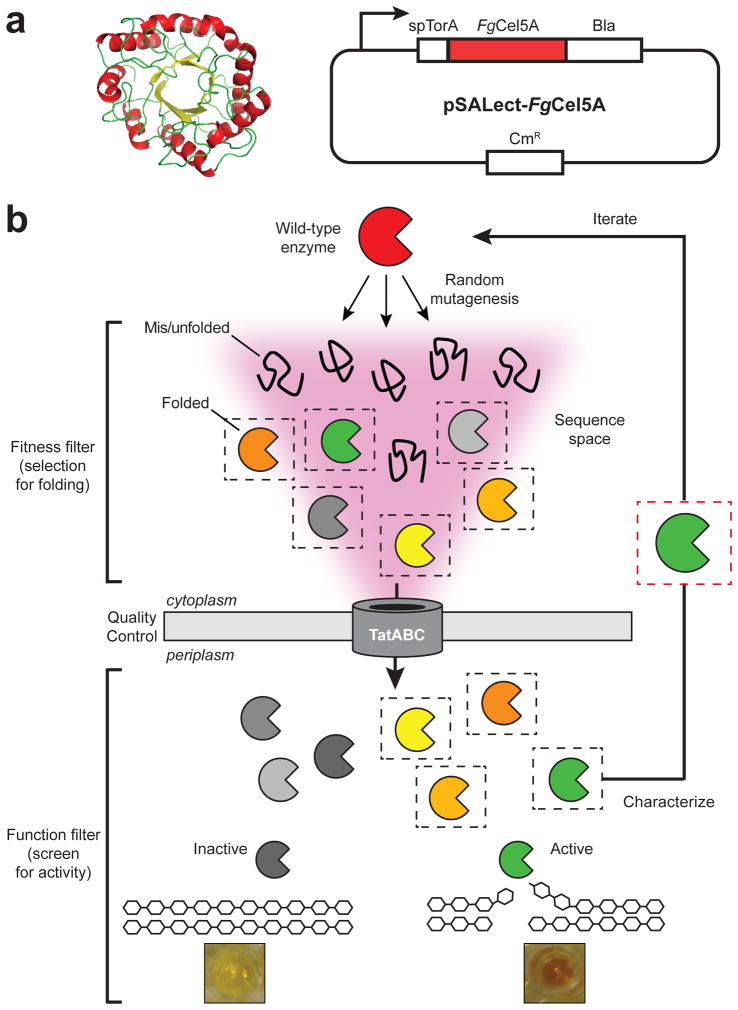
Figure 1. Optimizing protein production by directed evolution.
(a) Homology model of the FgCel5A CD (left) and plasmid pSALect-FgCel5A in which a Tat-dependent export signal (spTorA) directs the FgCel5A-Bla fusion to the periplasm of E. coli (right). (b) Schematic of the two-tiered directed evolution strategy. In the first tier, Tat QC-based genetic selection is applied, linking protein stability in vivo with resistance to β-lactam antibiotics. Since Bla must be in the periplasm to confer resistance to β-lactam antibiotics, only correctly folded fusion proteins that pass the Tat QC are capable of rendering cells drug resistant. Proteins that misfold and/or aggregate are not exported from the cytoplasm, and thus cells expressing these proteins are sensitive to β-lactam antibiotics. This selection allows for rapid isolation of well-folded FgCel5A library members while eliminating those that are poorly folded, thereby focusing the sequence space. In the second tier, an activity screen is imposed to ensure that the proteins that pass Tat QC retain high activity. For FgCel5A, this involves an enzymatic activity assay using the soluble cellulose substrate CMC. Following isolation of variants, the enzymes are characterized for soluble production and activity, and the fittest members are used for further iterations through the two-step selection and screen.