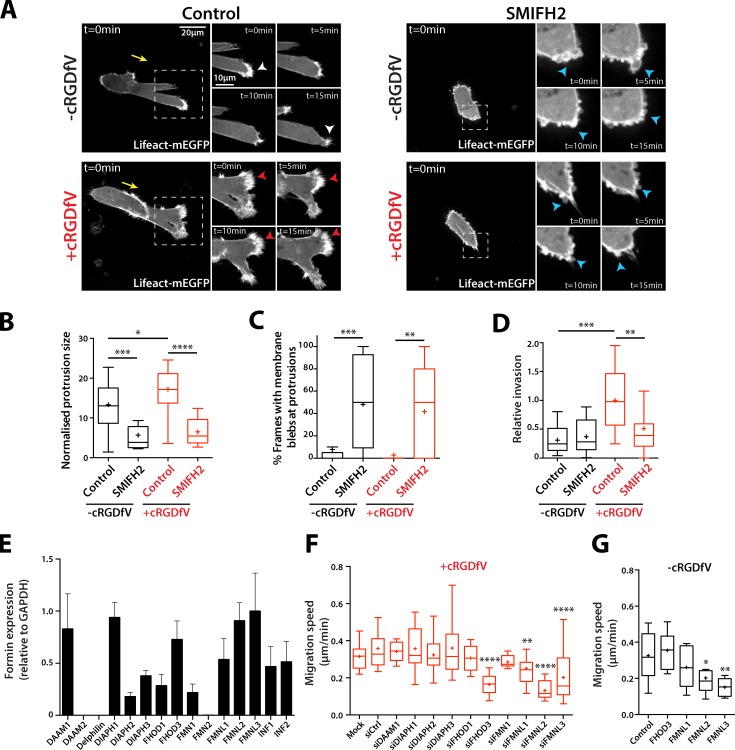
Figure 4.
Formin function is required for protrusion formation. (A) A2780 cells transiently transfected with Lifeact-mEGFP were seeded onto 3D CDM for 4 h, and treated with cRGDfV (2.5 µM, 2 h). Cells were treated with SMIFH2 (25 µM) or vehicle control immediately before imaging. Maximum z projections are shown. White arrowheads indicate lamellipodia-like protrusions; red arrowheads indicate actin spikes; blue arrowheads indicate blebs. Yellow arrows indicate direction of migration. The white boxed regions are enlarged on the right. (B) Normalized protrusion size of cells as in A. n = 12–20 cells per condition with n ≥ 25 frames quantified per cell. (C) Percentage of movie frames with bleb protrusions. n = 17 cells per condition. (D) A2780 cells were seeded into inverted invasion assays in the presence of cRGDfV (2.5 µM) where indicated. SMIFH2 (25 µM) and vehicle control were added 24 h after seeding, and invasion assays were imaged for quantification 48 h later. n = 15 per condition. (E) Formin expression in A2780 cells was measured by qRT-PCR. Gene expression is shown relative to GAPDH expression. n = 3. Error bars represent the SEM. (F and G) siRNA miniscreen of formins in A2780 cells. Migration speed on CDM in the presence of cRGDfV (2.5 µM; F) or under basal conditions (G) is shown. n = 10–30 cells per condition. All data represent at least three independent experiments. Statistical significance was evaluated using ANOVA/Tukey’s multiple comparison test. n.s., not significant; +, mean; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.