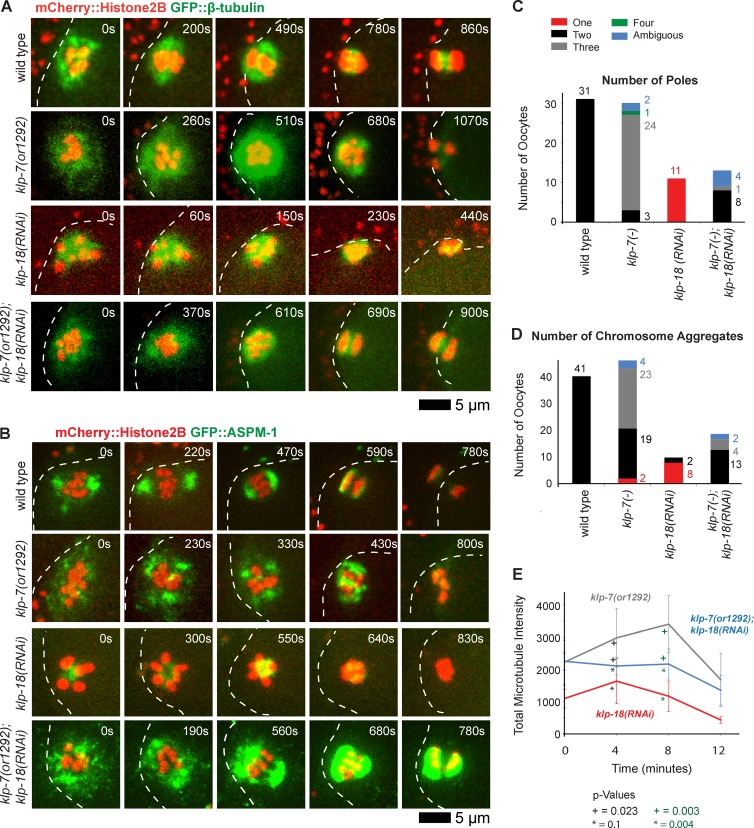
Figure 4.
Reducing klp-7 function partially restores oocyte meiotic spindle bipolarity in klp-18(−) mutants. (A and B) Time-lapse spinning disk confocal images during meiosis I of live wild-type and mutant embryos expressing either mCherry::Histone H2B and GFP::β-tubulin to mark chromosomes and microtubules, respectively (A), or mCherry::Histone H2B and GFP::ASPM-1 to mark chromosomes and spindle poles, respectively (B). White dashed lines indicate the oocyte plasma membrane, and times after ovulation are indicated in time-lapse sequence frames. (C and D) As described in Fig. 3, bar graphs indicate the number of GFP::ASPM-1 or GFP::MEI-1 foci in individual oocytes (C) and the number of segregating chromosome masses detected during anaphase from GFP::β-tubulin, GFP::ASPM-1, and GFP::MEI-1 strains expressing mCherry::Histone H2B (D) for the indicated genotypes. The legend (C) provides a color code for pole and aggregate numbers. The numbers from klp-7(RNAi) and klp-7(or1292ts) are merged; see Fig. S4 for the number of embryos analyzed for each genotype to score both pole and aggregate numbers. All data are from video micrographs of individual oocytes, each isolated from different worms in multiple experiments. The numbers of embryos scored as having each phenotype are adjacent to each bar. See Fig. S4 for bar graphs showing the results for each klp-7(−) genotype. (E) Integrated GFP::β-tubulin pixel intensity beginning at ovulation over time during meiosis I from strains of the indicated genotypes. Error bars depict the standard deviation at each time point. P-values comparing the microtubule signals in klp-18(−) and klp-7(−);klp-18(−) oocytes are indicated with asterisks. Note in B and in subsequent figures that GFP::ASPM-1 often appears to accumulate in higher levels in klp-7(−) mutants compared with klp-7(+) genotypes; although we have not quantified this difference, it may result from the increased microtubule accumulation in klp7(−) oocytes or from other influences of KLP-7 on ASPM-1.