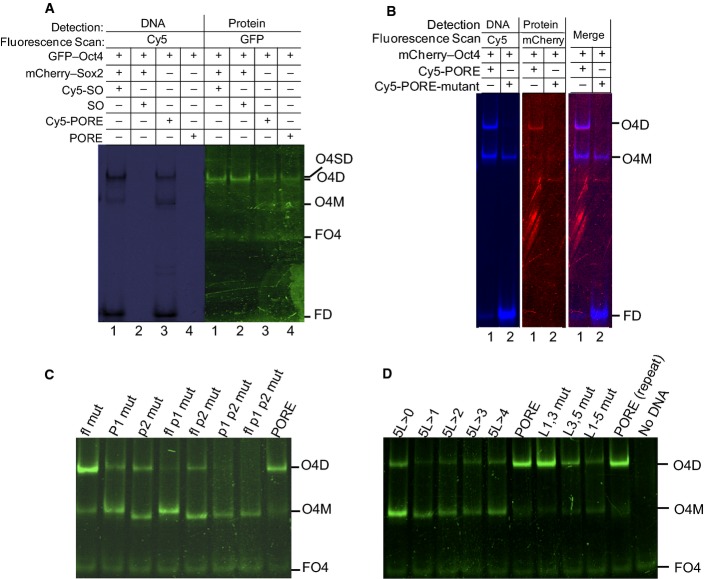
Validating FP-EMSA
- Homodimer formation was tested using DNA labelled with Cy5 or unlabelled as indicated. The presence of a Cy5 tag does not affect protein binding as dimerisation in lanes 2 and 4 is comparable with lanes 1 and 3.
- Homodimerisation is independent of the fluorescent protein tag. mCherry–Oct4 was tested instead of GFP-Oct4 with PORE-wt (lane 1) and PORE-mt (lane 2).
- Mutations inside the PORE motif affect homodimer formation, whereas mutations outside the PORE motif do not. The wild-type PORE is indicated; fl, P1 and P2 indicate oligonucleotides with mutations in the flanking sequence, palindromic repeat 1 and repeat 2, respectively.
- Effects of mutations of base pairs in the non-palindromic linker of the PORE motif (AAATG). 5L > 4-0 indicates sequential reduction in the size of the linker; other point mutation oligonucleotides are indicated.
Data information: O4D: GFP-Oct4 homodimer complex, O4SD: GFP-Oct4 and mCherry-Sox2 heterodimer complex, O4M: GFP-Oct4 monomer complex, FO4: free GFP-Oct4, and FD: free DNA. All oligonucleotide sequences are in Appendix Table S1. n = 3. Source data are available online for this figure.