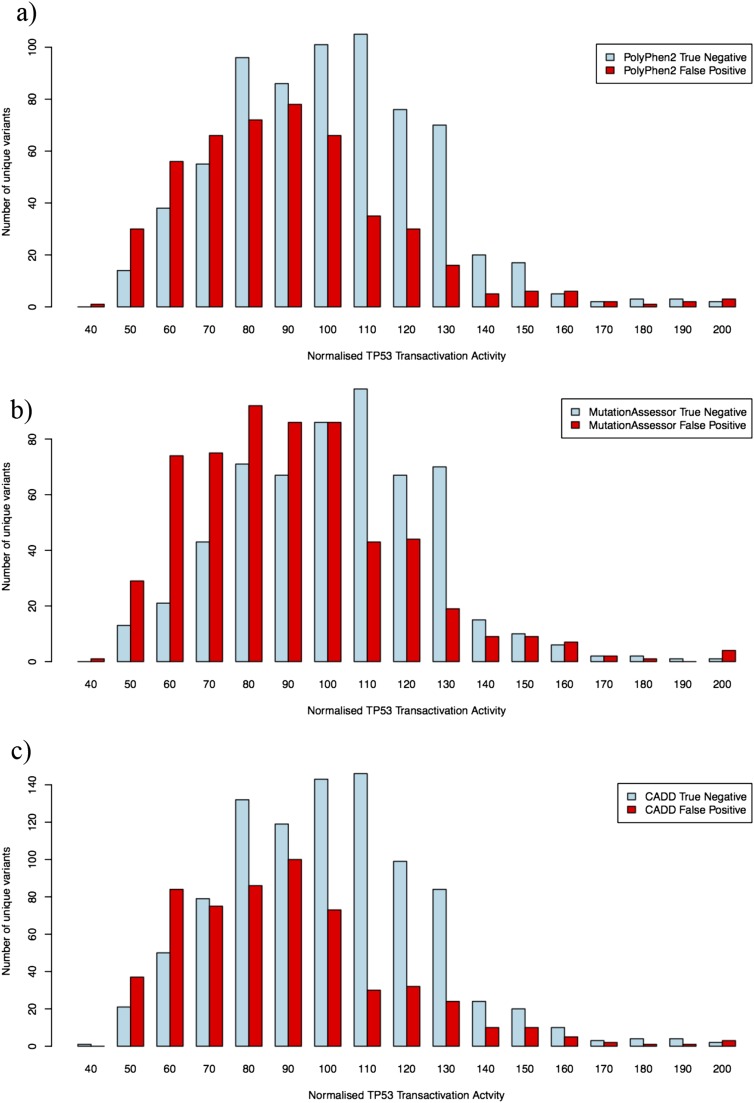
Fig. S2.
Distributions of residual TP53 transactivation activities of PolyPhen2-, MutationAssessor-, and CADD-assigned true-negative and false-positive mutations. Measured TP53 transactivation activity for true-negative (TN; blue) and false-positive (FP; red) categories of TP53 missense mutations identified in Fig. 2 A–C using inference scores calculated with (A) PolyPhen2, (B) MutationAssessor, and (C) CADD. For PolyPhen2, the mean normalized activity was 106.1 for the TN set and 94.6 for the FP set (Kolmogorov–Smirnov D = 0.2385, P = 2.454e−14; Wilcox W = 210,343.5, P = 3.541e−16). For MutationAssessor, the mean normalized activity was 104.1 for the TN set and 95.4 for the FP set (Kolmogorov–Smirnov D = 0.1792, P = 2.204e−10; Wilcox W = 327,022.5, P = 3.364e−12). For CADD, when intermediate values (score >5 and <20) are included with the TN category, the mean normalized activity was 105.3 for the TN set and 93.4 for the FP set (Kolmogorov–Smirnov D = 0.2401, P < 2.2e−16; Wilcox W = 190,543.5, P < 2.2e−16).