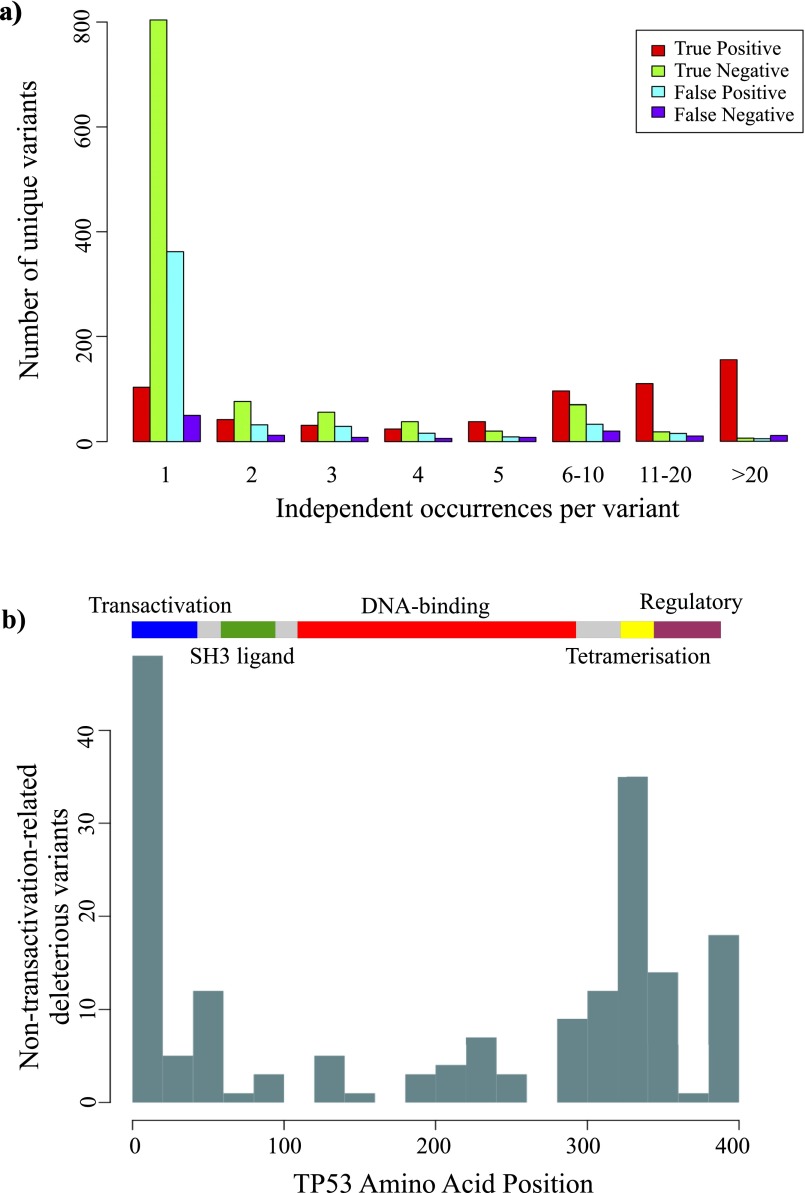
Fig. S3.
Incidence and location of apparent false-positive TP53 somatic mutations. (A) Unique TP53 somatic mutations found in human cancers, separated into categories as shown in Fig. 2D and ranked by the number of independent occurrences in separate cancer cases. Red, true positive; green, true negative; blue, false positive; purple, false negative. (B) The ideogram shows the major functional domains of the TP53 protein. Plotted against this are the number of false-positive mutations in 20 amino acid bins: from the set of all possible TP53 mutations, those that were predicted to be deleterious with Polyphen2 score >0.8 yet actually retained >40% of WT transactivation activity. Unlike most oncogenic TP53 mutations that occur in the DNA-binding domain (red), false-positive mutations appear concentrated in the extremities of the transactivation and tetramerization domains.