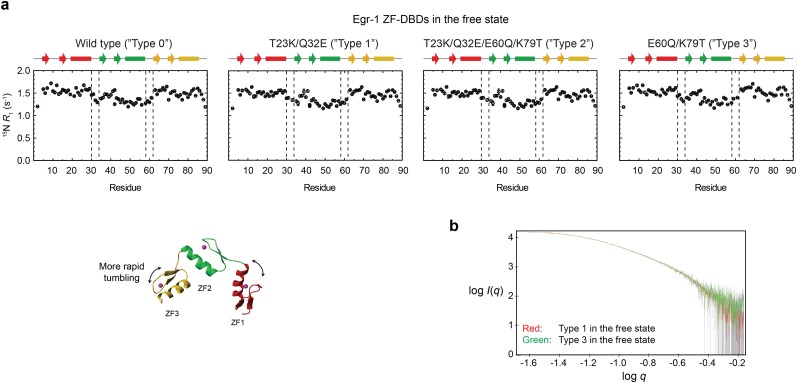
Fig. S3.
Impact of the T23K, Q32E, E60Q, and K79T mutations on the interdomain dynamics of the free Egr-1 ZF-DBD. (A) 15N longitudinal relaxation rates R1 measured for protein backbone amide groups of the Egr-1 ZF-DBDs. For all four constructs of types 0–3, ZF1 and ZF3 exhibited larger R1 rates than ZF2, presumably because of less restricted domain motions (i.e., one linkage vs. two linkages). It should be noted that the 15N R1 profiles are similar for all of the four proteins in the free state. (B) SAXS data for the type 1 (red) and type 3 (green) mutant proteins in the free state. The SAXS data were collected with a Rigaku BioSAXS-1000 instrument using 4 mg/mL protein in the same buffer as that used for the NMR experiments. Intensity I is plotted as a function of the scattering vector q with logarithmic scales for both axes. The SAXS data for these proteins were statistically identical. These NMR and SAXS data indicate that the mutations do not significantly impact the interdomain dynamics of the Egr-1 ZF-DBD in the free state.