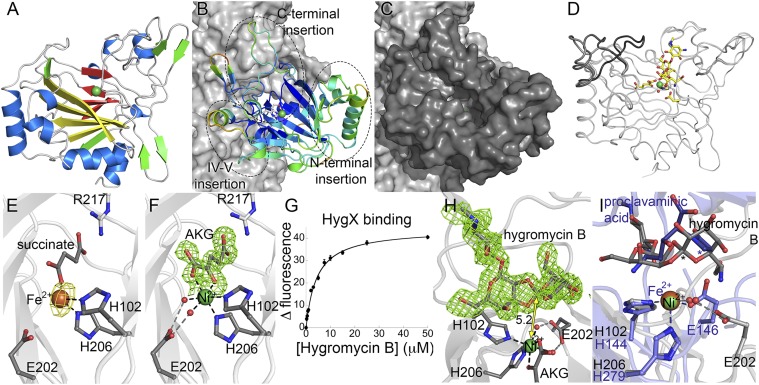
Fig. 2.
HygX structure. (A) HygX with major strands in yellow and minor strands in red. Ni2+and Fe2+ are green and orange spheres. (B) HygX A chain colored by crystallographic temperature factors where cool colors indicate low B-factors. Loop insertions are highlighted. (C) Surface representation in the same orientation as B. (D) Superposition of four chains of the HygX-AKG complex showing loop movement, with hygromycin B shown for reference. (E) Iron coordination with anomalous difference density in yellow and contoured at 5σ. (F) Active site metal coordination of HygX. |FO| − |FC| difference density is contoured around AKG at 2.5 σ, calculated before inclusion of AKG in the model. (G) Tryptophan fluorescence at 350 nm of 0.5 μM HygX, 0.05 mM NiCl2, 0.05 mM AKG, and varying concentrations of hygromycin B (Kd, 3.4 ± 0.5 μM). (H) 3σ |FO| − |FC| difference density calculated before hygromycin B placement. One possible site of hydrogen atom abstraction is 5.2 Å from the metal center. (I) HygX–AKG–hygromycin B product complex (in gray) aligned with the clavaminate synthase–AKG–proclavaminate substrate complex (blue; PDB ID code 1DRT). Asterisks mark sites of hydrogen atom abstraction.