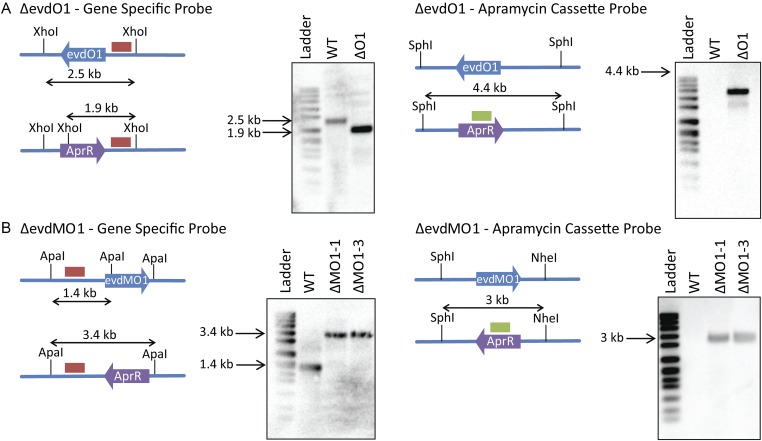
Fig. S8.
Southern hybridization of targeted deletion mutants verifying a double crossover event. (A) Southern blot analysis of ΔevdO1. Diagrams depict the relative shifts expected for replacement of evdO1 with the apramycin cassette. XhoI and SphI are restriction endonucleases used to cleave the genomic DNA into predictably sized fragments. Blots show predicted shifts were observed experimentally, thus confirming the double crossover. Ladder is DNA molecular-weight marker VII, DIG-labeled (product no. 11669940910; Roche Life Sciences). WT is wild-type M. carbonacea var aurantiaca, and ΔO1 is the ΔevdO1 knockout strain. (B) Southern blot analysis of ΔevdMO1. Diagrams depict the relative shifts expected for replacement of evdMO1 with the apramycin cassette. ApaI, KpnI, and NheI are restriction endonucleases used to cleave the genomic DNA into predictably sized fragments. Blots show predicted shifts were observed experimentally, thus confirming the double crossover. Ladder is DNA molecular-weight marker VII, DIG-labeled (product no. 11669940910; Roche Life Sciences). WT is wild-type M. carbonacea var aurantiaca, and ΔMO1-1 and ΔMO1-3 are ΔevdMO1 knockout strains.