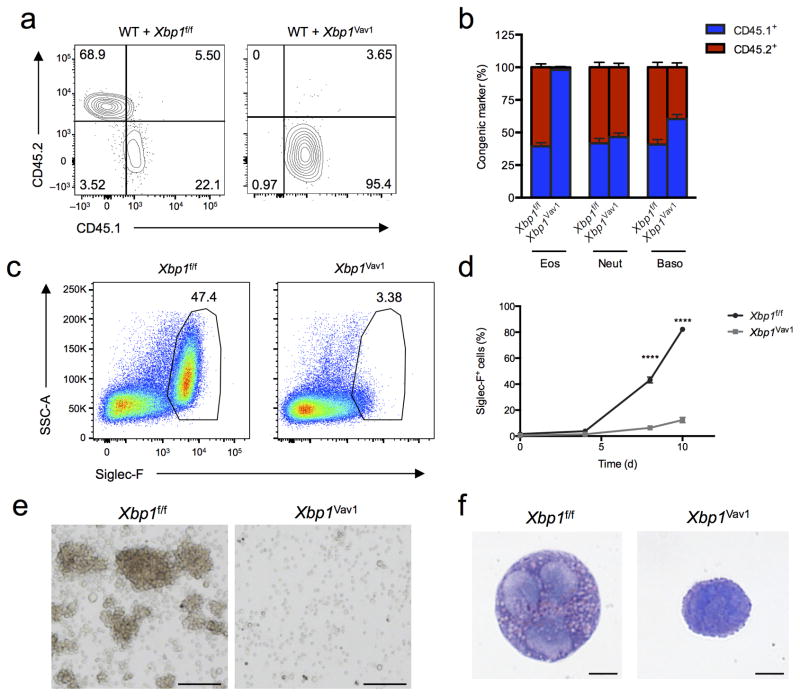
Figure 3.
XBP1 is a cell-intrinsic requirement for eosinophil development. (a) Flow cytometry assessing expression of the congenic markers CD45.2 (Xbp1f/f or Xbp1Vav1) and CD45.1 (wild type) on peripheral blood eosinophils from chimeras generated with a mixture of wild-type bone marrow plus either Xbp1f/f bone marrow (WT + Xbp1f/f) or Xbp1Vav1 bone marrow (WT + Xbp1Vav1). Numbers in quadrants indicate percent cells in each. (b) Reconstitution efficiency of eosinophils, neutrophils (Neut) and basophils (Baso) in blood from mixed-bone marrow chimeras as in a (n = 6 host mice per chimera type). (c) Flow cytometry analyzing Siglec-F expression Xbp1f/f and Xbp1Vav1 BMDEs at day 8 of culture, gated on DAPI− singlets. Numbers adjacent to outlined areas indicate percent Siglec-F+ cells. (d) Frequency of Siglec-F+ cells among Xbp1f/f and Xbp1Vav1 BMDEs at days 0, 4, 8 and 10 of culture (n = 3 independent cultures per genotype). (e) Microscopy of hematopoietic colonies from Xbp1f/f and Xbp1Vav1 BMDEs at day 8 of culture. Scale bars, 100 μm. (f) Microscopy of cytospin analysis of DAPI−Siglec-F+ singlets sorted from Xbp1f/f and Xbp1Vav1 BMDEs at day 8 of culture. Scale bars, 5 μm. *P < 0.0001 (two-way analysis of variance (ANOVA) with the Šidák correction for multiple comparisons). Data are from two independent experiments with six mice per group (a,b; mean and s.e.m. in b) or more than three independent experiments with at least three mice per group (c–f; mean and s.e.m. in d).