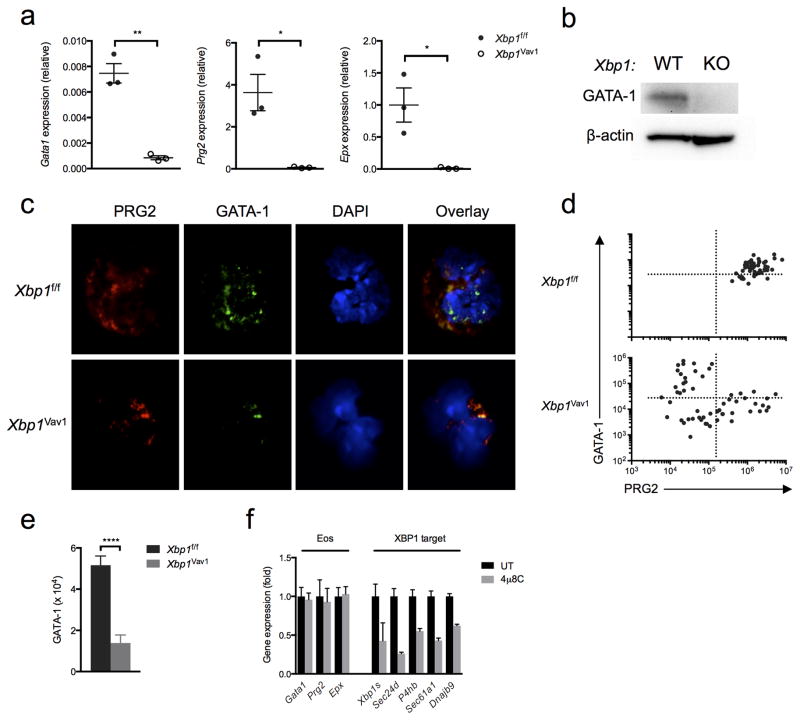
Figure 8.
Xbp1 deficiency prevents the accumulation of GATA-1 in eosinophils through an indirect mechanism. (a) Quantitative PCR analysis of Gata1, Prg2 and Epx in sorted DAPI−Siglec-F+ BMDEs from Xbp1f/f and Xbp1Vav1 mice (n = 3 independent cultures per genotype), assessed at day 8 of culture; results were normalized to those of Actb. (b) Immunoblot analysis of GATA-1 and β-actin (loading control) in Xbp1f/f and Xbp1Vav1 BMDEs at day 8 of culture. (c) Immunofluorescence microscopy of developing eosinophils among Xbp1f/f and Xbp1Vav1 BMDEs at day 8 of culture, stained for PRG2, GATA-1 and DAPI. (d) Scatter plots of PRG2 fluorescence versus GATA-1 fluorescence in Xbp1f/f BMDEs (n = 50) and Xbp1Vav1 BMDEs (n = 56) at day 8 of culture. (e) Staining intensity of GATA-1 in developing PRG2+ eosinophils among Xbp1f/f BMDEs (n = 47) and Xbp1Vav1 BMDEs (n = 30) at day 8 of culture. (f) Quantitative PCR analysis of the canonical eosinophil genes Gata1, Prg2 and Epx, as well as Xbp1s and the XBP1 target genes Sec24d, P4hb, Sec61a1 and Dnajb9, in BMDE cultures treated for 8 h with the vehicle dimethyl sulfoxide (DMSO) or 4μ8C (20 μM) (n = 3 independent cultures per condition); results were normalized to those of Actb and are presented relative to those of vehicle-treated cells. *P < 0.05 and **P < 0.01 and ***P < 0.0001 (Student’s t-test). Data are representative of at least two independent experiments (a,f; mean and s.e.m.), two independent experiments (b) or two experiments with three independent biological replicates per genotype (c) or are from one experiment with three biological replicates pooled per genotype (d,e; mean and s.e.m. in e).