Figure 8.
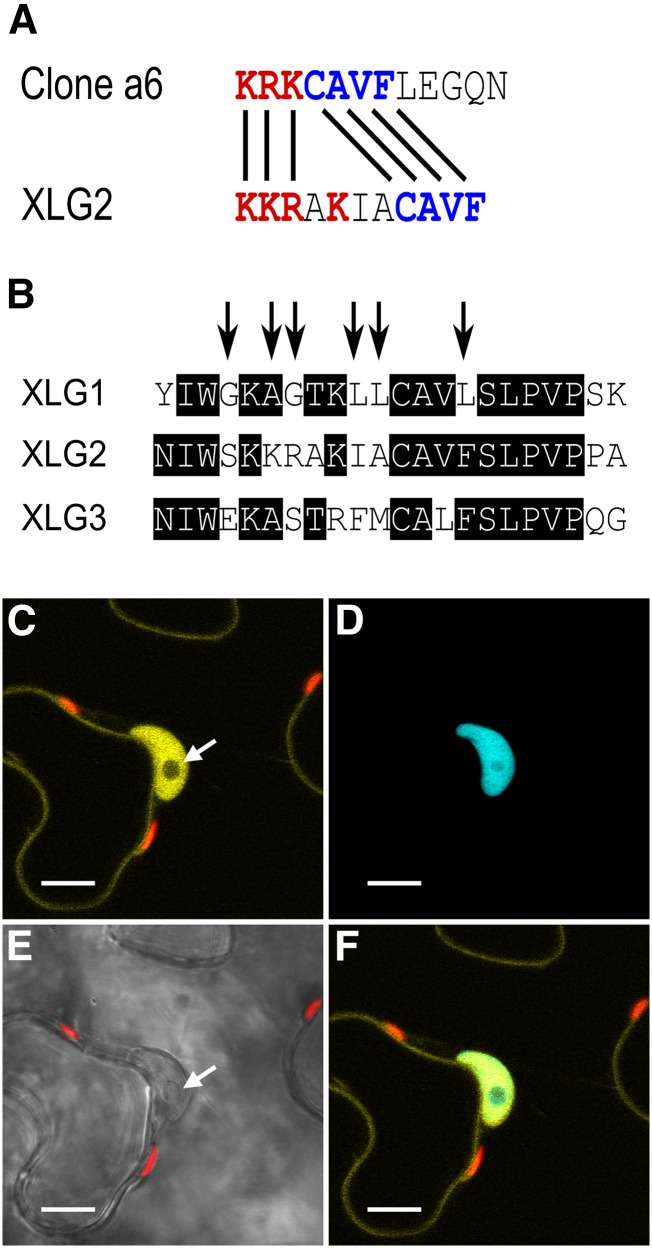
XLG2 has a functional NLS. A, The XLG2 NLS has a similar basic-positive patch of residues (red) and a CAVF motif (blue) identical to the empirically validated clone a6 sequence (Kosugi et al., 2009). B, The XLG2 NLS resides in a semiconserved stretch of sequence in the XLG family starting at XLG2 residue 424, and the arrows show the XLG1 residues mutated to mirror XLG2 to create the XLG1m1 construct. XLG1m1:mVenus fusions localized to the nucleus in N. benthamiana leaves, demonstrating that the XLG2 NLS is a functional regulatory domain. (Compare with XLG1:mVenus localization in Figure 7, A and B.) C to F, Agroinfiltration of XLG1m1:mVenus into N. benthamiana leaves at an OD600 of 0.0075 to 0.01 and imaged 48 h later at 63× magnification. C, XLG1m1:mVenus localizes to the interior of the nucleus and at the plasma membrane; the nucleolus (white arrow) does not appreciably accrue XLG1m1. D, The nucleus is specifically marked by mTq2:histone 2B. E, The nucleus and nucleolus (white arrow) are clearly visible in the bright-field channel. F, XLG1m1:mVenus and mTq2:histone 2B colocalization is specific to the nucleus. Yellow indicates mVenus, blue indicates mTq2, and red indicates chlorophyll autofluorescence. Bars = 10 μm.