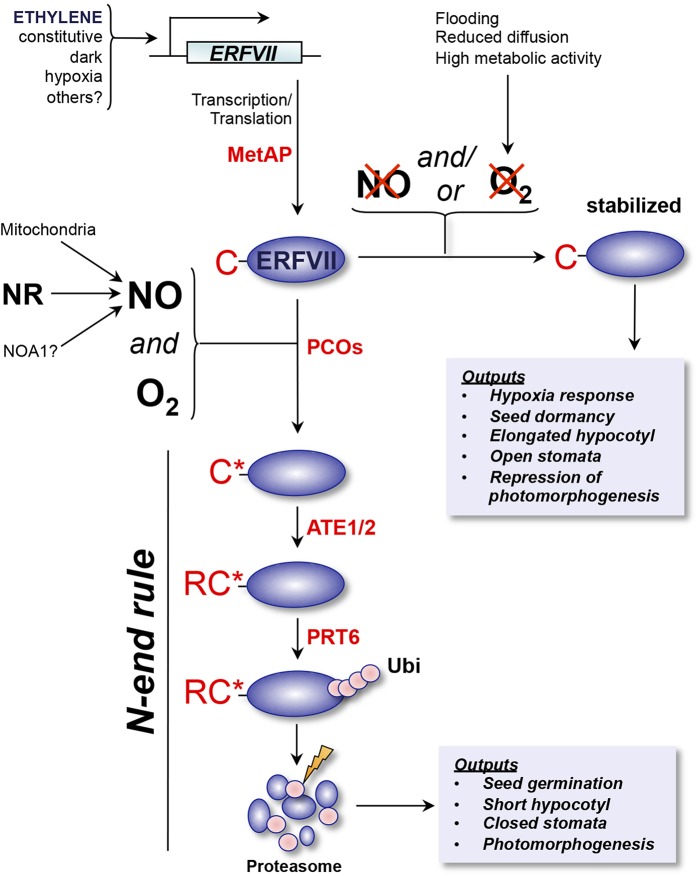
Figure 2.
Oxygen (O2) and nitric oxide (NO) signal transduction via N-end rule regulation of ERFVII stability. Arabidopsis ERFVII transcription factors are transcribed either constitutively and/or in response to several upstream signals, including ethylene. MET AMINOPEPTIDASE (MetAP) enzymes cleave Nt-Met cotranslationally to reveal Nt-Cys (C). In the presence of oxygen and NO (which may be derived from a variety of cellular sources: NR, nitrate reductase; NOA1, nitric oxide associated protein1), Nt-Cys is oxidized to Cys-sulfinic or Cys-sulfonic acid (C*). This oxidation is facilitated by PLANT CYS OXIDASE (PCO) enzymes (Weits et al., 2014). Oxidized Nt-Cys is then arginylated (R) by ARGINYL tRNA TRANSFERASES (ATE1/2). Nt-Arg-ERFVII is recognized by the N-end rule E3 ligase (N-recognin) PROTEOLYSIS6 (PRT6), which targets the protein for proteasomal degradation via polyubiquitination (Ubi; Gibbs et al., 2011; Licausi et al., 2011). ERFVII degradation initiates germination, inhibits hypocotyl growth, stimulates stomatal closure, represses the hypoxia transcriptional response, and promotes photomorphogenesis. In conditions in which either oxygen or NO (or both) become limiting, Nt-Cys oxidation is inhibited, and ERFVIIs are stabilized (Gibbs et al., 2014b). Stable ERFVIIs promote anaerobic gene expression, maintain seed dormancy and open stomata, promote hypocotyl growth, and repress photomorphogenesis. Regulation of ERFVII stability by the N-end rule pathway therefore controls the homeostatic response to oxygen and NO availability.