Figure 5.
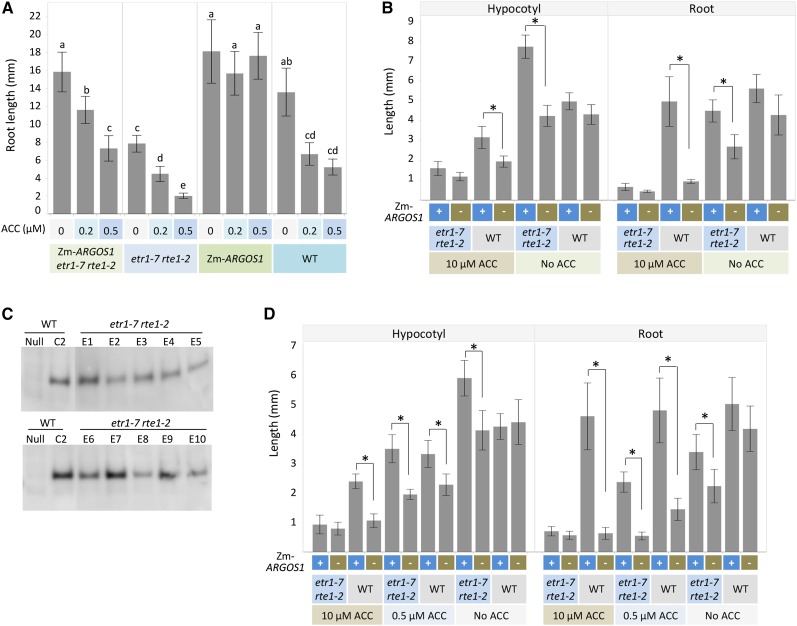
Overexpression of ZmARGOS1 in the Arabidopsis etr1-7 rte1-2 double mutant. A, Root lengths in the 35S:ZmARGOS1 etr1-7 rte1-2 Arabidopsis plants compared with the etr1-7 rte1-2 double mutant. The 35S:ZmARGOS1 etr1-7 rte1-2 plant was generated by crossing the ZmARGOS1 event 2B3 and the etr1-7 rte1-2 double mutant followed by self-pollination. Plants were grown for 5 d in agar that contained one-half-strength Murashige and Skoog medium with 0, 0.2, or 0.5 µm ACC and were set vertically in a growth chamber under a regime of 16 h of light (approximately 120 mE m−2 s−1) at 24°C and 8 h of darkness at 23°C. The root lengths of the 35S:ZmARGOS1 plants in the wild-type (WT) background showed no significant difference under 0, 0.2, and 0.5 µm ACC (P > 0.05, ANOVA), but the 35S:ZmARGOS1 etr1-7 rte1-2 plants were significantly different (P < 0.05, ANOVA). Significant differences are denoted by different letters. Data are means ± sd; n = 10 to 20. B, Hypocotyl and root lengths of 3-d-old, etiolated Arabidopsis seedlings are presented for the 35S:ZmARGOS1 etr1-7 rte1-2 plants and the etr1-7 rte1-2 double mutant. The 35S:ZmARGOS1 etr1-7 rte1-2 plant was generated by crossing the ZmARGOS1 event 2B3 and the etr1-7 rte1-2 double mutant followed by self-pollination. The triple response assay was conducted in the presence and absence of 10 µm ACC in the dark. The wild-type Arabidopsis transgenic plants carrying 35S:ZmARGOS1 and nontransgenic wild-type plants served as controls. Error bars indicate sd; n = 10. Student’s t test was performed to compare the plants with or without the ZmARGOS1 transgene. *, P < 0.01. C, Immunoblot analysis of ZmARGOS1 overexpression in the etr1-7 rte1-2 Arabidopsis mutant plants. A FLAG-HA epitope-tagged ZmARGOS1 was overexpressed in the double mutant (etr1-7 rte1-2) or the wild-type background under the control of the cauliflower mosaic virus 35S promoter. Ten events (E1–E10) are shown. Event C2 of 35S:ZmARGOS1-FLAG-HA in the wild-type background served as a positive control. Western-blot analysis was performed with anti-FLAG antibodies. D, Hypocotyl and root lengths of 3-d-old, etiolated Arabidopsis seedlings are presented for the 35S:ZmARGOS1 etr1-7 rte1-2 plants and the etr1-7 rte1-2 double mutant. The 35S:ZmARGOS1 etr1-7 rte1-2 plant was generated by transforming the etr1-7 rte1-2 double mutant plants with 35S:ZmARGOS1-FLAG-HA. The triple response assay was conducted with four independent events (E1, E4, E7, and E9 in C) in the presence of 0, 0.5, or 10 µm ACC in the dark. Results from event E1 are shown. The wild-type Arabidopsis transgenic plants carrying 35S:ZmARGOS1 and nontransgenic wild-type plants served as controls. Error bars indicate sd; n = 10. Student’s t test was performed to compare the plants with or without the ZmARGOS1 transgene. *, P < 0.01.