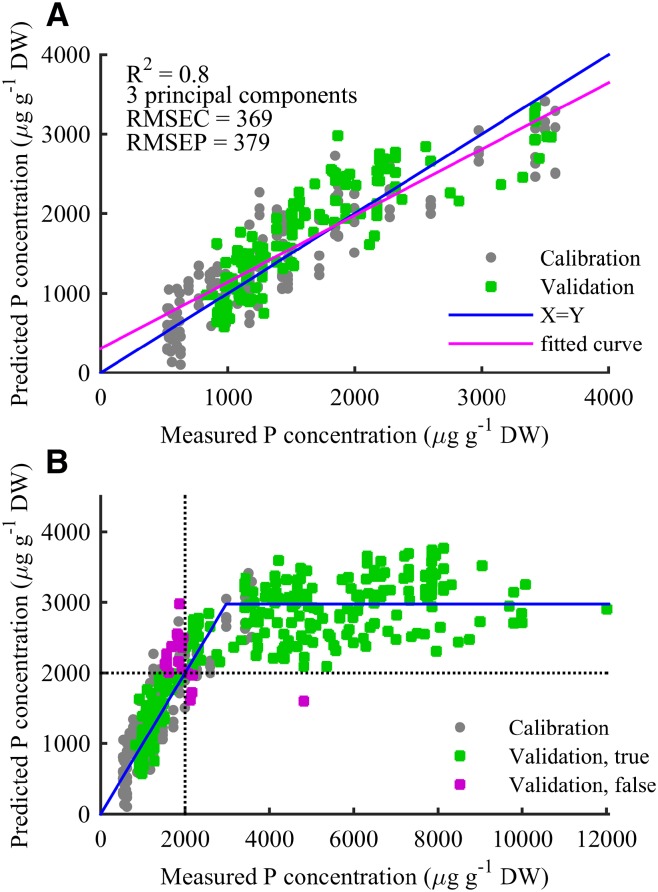
Figure 4.
Predicted versus measured barley leaf P concentrations based on a PLS model with three principal components. A, PLS prediction in the calibration range. Gray circles indicate calibration samples, and red circles indicate validation samples. The blue curve indicates the optimal Y = X line, and the red curve represents best linear fit to data (R2 = 0.8). The root-mean-squared error for the 159 calibration samples (root-mean-squared error of calibration [RMSEC] = 369) is seen to be very equivalent to the root-mean-squared error for the 131 independent test samples (root-mean-squared error of prediction [RMSEP] = 379). Together with the low number of principal components used, this shows that the model is not a result of overfitting. B, Predicted versus measured P concentrations for all 448 OJIP transients. The blue curve represents the optimal fit of a Y = X line intersecting a constant line at 2,975 µg g–1 DW. The two dotted lines indicate 2,000 µg g–1 DW, and coloring indicates whether the PLS model predicts samples correctly according to this threshold. Two hundred sixty-nine of 291 leaves (92%) from the test set were correctly classified as above or below the 2,000 µg g–1 DW threshold, and the misclassified leaves position close to the threshold except for one measurement.