Figure 10.
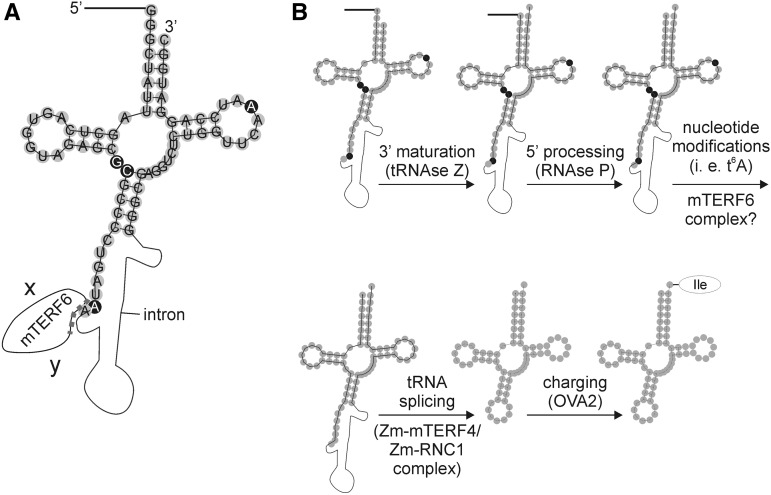
Hypothetical maturation steps of Arabidopsis tRNAIle(GAU) (trnI.2). A, Schematic representation of Arabidopsis plastid pre-trnI.2. The mature trnI.2 was drawn with the RNAfold Webserver (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi), and pre-tRNA properties were added manually. Filled gray circles represent exon nucleotides, and the black line and the thin black curve represent the 5′ leader and the intron, respectively. Nucleotides that are posttranscriptionally modified in human mitochondrial tRNAIle(GAU) (Suzuki and Suzuki, 2014) are indicated by white letters in black circles. In human mitochondrial tRNAIle(GAU), nucleotide A-37 is threonyl-carbamoylated, leading to t6A. The identified mTERF6 binding site (marked with the dotted line) spans the last nucleotide (A-38) of the first exon and the very beginning of the trnI.2 intron. The letters x and y symbolize yet unidentified interaction partners of mTERF6. B, Schematic representation of the tentative maturation process. Precursor tRNAs are transcribed with additional sequences at the 5′ and 3′ ends of the tRNA (not shown here), and some tRNA genes contain introns, as is the case for trnI.2. The tRNA has to undergo several maturation steps before it is functional and can be charged with its amino acid. The order of tRNA processing events is derived from tRNA maturation steps in yeast (for review, see Hopper et al., 2010) and defining the maturation status of M. kandleri tRNAs with the help of RNA sequencing data (Su et al., 2013). The hypothetical order of observed tRNA processing events is as follows: (1) 3′ processing, (2) 5′ processing, (3) nucleotide modifications, (4) splicing, and (5) charging with its amino acid. However, the exact chronological order might deviate for various tRNAs. ZmmTERF4 and ZmRNC1 are proteins that are required for tRNAIle(GAU) intron splicing in maize.