Figure 6. Effect of the amplitude threshold setting in the spot detection algorithm.
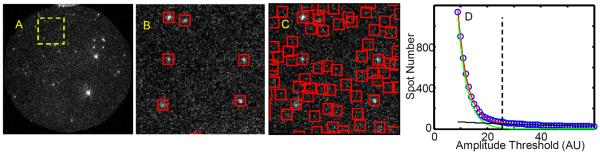
(A) Circular 65 μM diameter binder channel image. There are two bright spots due to 40 nM fluorescent bead fiducial markers and ~70 dimmer spots arising from Cy3B-GreB binder molecules bound to transcription elongation complex targets. (B, C) Magnified view of the 13.2 × 13.2 μm region enclosed by the dotted yellow line in (A). The spot detection algorithm detects 6 spots (red squares in (B)) at the high spot amplitude threshold equal to 25 and 60 spots (C) at the low threshold equal to 9. Images in (A–C) are reproduced at high contrast to emphasize image noise. (D) The number of detected spots (blue) for the field of view shown in (A) varies with the amplitude threshold setting. At low amplitude settings (i.e., <20) the algorithm identifies image noise features as spots, resulting in an excessive number of detected spots. The data are fit with a bi-exponential function (red) consisting of one term approximating the number of false positive spots due to image noise (green) and a second term approximating the number of true binder spots (black). The fits are used to calculate a threshold (dashed line) for which there is an estimated 50% probability of having no noise-induced spots detected within 1 pixel of a single DNA location during the 4000 frame duration experiment.
