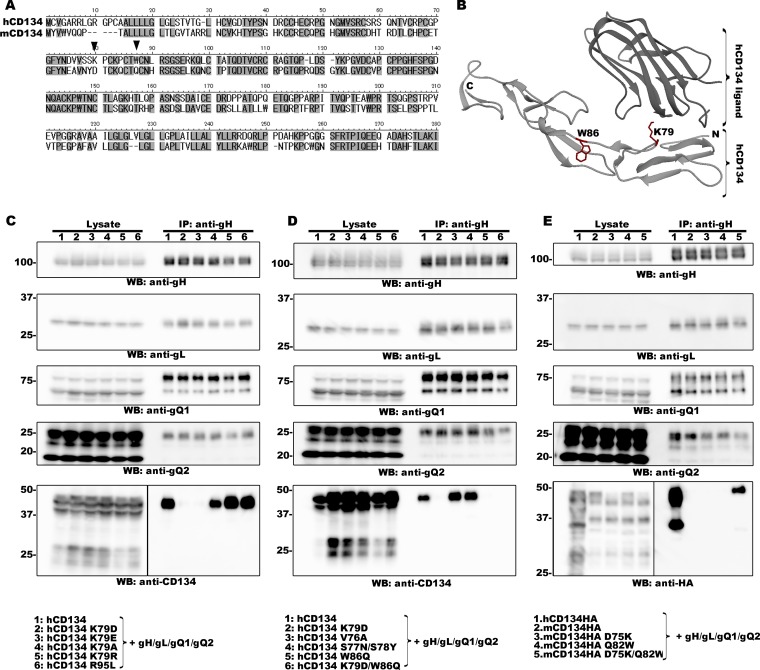
FIG 1.
Identification of two amino acid residues within CD134 that are required for HHV-6B ligand binding. (A) Amino acid sequence alignment of human and murine CD134. The two amino acid residues of interest are indicated by black triangles. (B) Crystal structure of hCD134 and its ligand (PBD accession number 2HEV). (Bottom panel) Crystal structure of hCD134 (K79 and W86 are shown with their side chains in red); (upper panel) ligand for hCD134. The structure drawing was made by using UCSF Chimera software (23; www.cgl.ucsf.edu/chimera). (C, D, and E) 293T cells were transfected with plasmids expressing the proteins (indicated by a number at the top of each lane and identified at the bottom of the figure) by using Lipofectamine 2000 (Invitrogen). The transfected cells were harvested at day 2 posttransfection and lysed with TNE buffer (10 mM Tris-HCl [pH 7.8], 0.15 M NaCl, 1 mM EDTA, 1% NP-40 [Nacalai Tesque]) containing a cocktail of protease inhibitors (Sigma). The lysates were subjected to ultracentrifugation prior to immunoprecipitation (IP) with an anti-gH antibody. Expression of each protein (Lysate) and the proteins precipitated from the lysates (IP: anti-gH) were confirmed by Western blotting (WB). Numbers at the left of the gels are molecular masses (in kilodaltons).